Special topic: Metagenomics - Genome Sciences
Special topic: Metagenomics - Genome Sciences
Special topic: Metagenomics - Genome Sciences
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
grouping shows that our current GOS sampling methodology<br />
will not cover all protein families, and perhaps misses some<br />
protein families Rate that are of exclusive protein to higher discovery eukaryotes. The<br />
large section of clusters that include all three groupings<br />
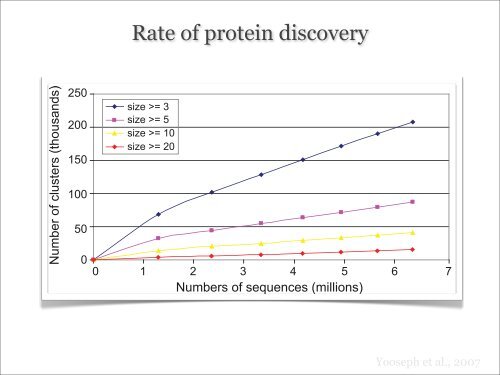
uences Are Added<br />
e y-axis denotes the number of clusters (in thousands). Seven datasets with increasing<br />
d in the text. The blue curve shows the number of core sets of size 3 for the seven<br />
own. Linear regression gives slopes 0.027 (R 2 ¼ 0.999), 0.011 (R 2 ¼ 0.999), 0.0053 (R 2 ¼<br />
nd size 20, respectively.<br />
only sequenc<br />
members of k<br />
clustering pa<br />
detected by a<br />
Figure 2. Rate of Discovery of Clusters as (Nonredundant) Sequences Are Added<br />
The x-axis denotes the number of sequences (in millions) and the y-axis denotes the number<br />
numbers of (nonredundant) sequences are chosen as described in the text. The blue curve<br />
datasets. Curves for core set sizes 5, 10, and 20 are also shown. Linear regression gives<br />
0.999), and 0.0024 (R 2 0437<br />
March 2007 | Volume 5 | Issue 3 | e16<br />
¼ 0.996) for size 3, size 5, size 10, and size Yooseph 20, et respectively. al., 2007