2009 Scientific Report
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Van Andel Research Institute<br />
<strong>Scientific</strong> <strong>Report</strong> <strong>2009</strong>
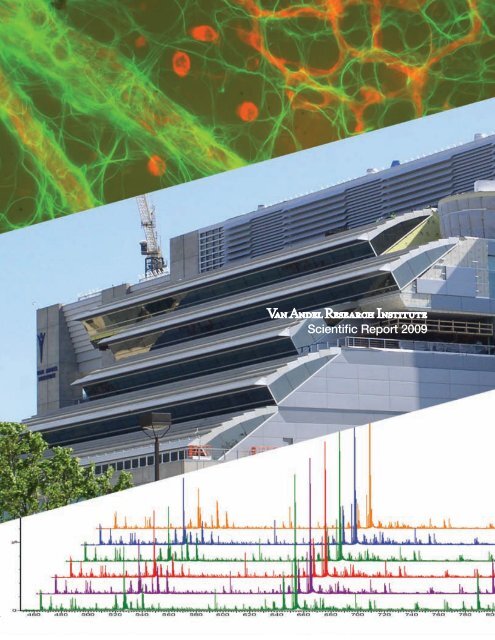
Cover photo: The micrograph at the top of the cover shows astrocytes in the murine retina (see<br />
p. 46); photo by Jennifer Bromberg-White. The center photo shows the VARI Phase II construction<br />
from the west as it appeared in May <strong>2009</strong>; photo by David Nadziejka. The lower figure is a<br />
series of liquid chromatography–mass spectrometry (LC-MS) spectrographs of complex protein<br />
samples (see p. 34); graphs courtesy of Greg Cavey.
VARI | <strong>2009</strong><br />
Van Andel Research Institute <strong>Scientific</strong> <strong>Report</strong> <strong>2009</strong><br />
Culture of prostate epithelial cells.<br />
Confocal microscopic image of prostate epithelial cell (PEC) acini. Cells were cultured on Matrigel for 15 days and immunostained with antibodies<br />
against integrin beta 1 (green) and laminin 5 (red) to delineate the interface of the cell and the secreted basement membrane. Blue is from<br />
Hoechst staining of DNA in the nuclei. The equatorial cross section shows that this is a hollow structure, recapitulating the in vivo characteristics<br />
of the prostate gland. Three-dimensional culture may provide a more physiologically relevant approach than traditional two-dimensional<br />
cultures for studying the regulation of survival pathways, cellular architecture, and other cellular processes ex vivo.<br />
Photo by Laura Lamb of the Miranti lab.
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Copyright <strong>2009</strong> by the Van Andel Institute; all rights reserved.<br />
Van Andel Institute, 333 Bostwick Avenue, N.E.,<br />
Grand Rapids, Michigan 49503, U.S.A.<br />
ii
VARI | <strong>2009</strong><br />
Director’s Introduction 1<br />
George F. Vande Woude, Ph.D.<br />
Laboratory <strong>Report</strong>s 7<br />
Arthur S. Alberts, Ph.D.<br />
Cell Structure and Signal Integration 8<br />
Brian Cao, M.D.<br />
Antibody Technology 11<br />
Gregory S. Cavey, B.S.<br />
Mass Spectrometry and Proteomics 14<br />
Nicholas S. Duesbery, Ph.D.<br />
Cancer and Developmental Cell Biology 17<br />
Bryn Eagleson, B.S., RLATG<br />
Vivarium and Transgenics Program 20<br />
Kyle A. Furge, Ph.D.<br />
Computational Biology 23<br />
Brian B. Haab, Ph.D.<br />
Cancer Immunodiagnostics 26<br />
Table of Contents<br />
Jeffrey P. MacKeigan, Ph.D.<br />
Systems Biology 30<br />
Cindy K. Miranti, Ph.D.<br />
Integrin Signaling and Tumorigenesis 35<br />
James H. Resau, Ph.D.<br />
Division of Quantitative Sciences<br />
Analytical, Cellular, and Molecular Microscopy<br />
Microarray Technology<br />
Molecular Epidemiology 39<br />
Pamela J. Swiatek, Ph.D., M.B.A.<br />
Germline Modification and Cytogenetics 43<br />
Bin T. Teh, M.D., Ph.D.<br />
Cancer Genetics 47<br />
Steven J. Triezenberg, Ph.D.<br />
Transcriptional Regulation 51<br />
George F. Vande Woude, Ph.D.<br />
Molecular Oncology 55<br />
Craig P. Webb, Ph.D.<br />
Program for Translational Medicine 59<br />
Michael Weinreich, Ph.D.<br />
Chromosome Replication 64<br />
Bart O. Williams, Ph.D.<br />
Cell Signaling and Carcinogenesis 68<br />
H. Eric Xu, Ph.D.<br />
Structural Sciences 72<br />
iii
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Daniel Nathans Memorial Award 76<br />
Dennis J. Slamon, M.D., Ph.D., and Genentech, Inc.<br />
Postdoctoral Fellowship Program 78<br />
List of Fellows<br />
Student Programs 80<br />
Grand Rapids Area Pre-College Engineering Program<br />
Summer Student Internship Program<br />
Han-Mo Koo Memorial Seminar Series 84<br />
2008 | <strong>2009</strong> Seminars<br />
Van Andel Research Institute Organization 89<br />
Boards<br />
Office of the Director<br />
VAI Administrative Organization<br />
iv
VARI | <strong>2009</strong><br />
Director’s Introduction<br />
1
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
George F. Vande Woude<br />
Director’s Introduction<br />
The year 2008 was extraordinary for the Van Andel Research Institute, with many of the highlights coming late. On October 1,<br />
the Institute celebrated the “topping out” of our Phase II building project with the installation of the topmost beam of the<br />
structure. Van Andel staff had a chance to sign the beam before the ceremony and to watch as it was swung aloft and bolted<br />
into place.<br />
In December, we celebrated the year’s recipients of the Daniel Nathans Award. VARI was pleased to honor Dr. Dennis J.<br />
Slamon, of the University of California, Los Angeles, and the firm Genentech for their roles in the development of the cancer<br />
therapeutic Herceptin. Dr. Slamon gave a <strong>Scientific</strong> Lecture entitled “Molecular diversity of human breast cancer: clinical and<br />
therapeutic implications”. Dr. Arthur D. Levinson accepted for the many members of Genentech who moved forward the first of<br />
the modern-era drugs that target known cancer genes. Dr. Levinson, CEO of Genentech, was one of its champions. He gave<br />
a <strong>Scientific</strong> Lecture entitled “Herceptin: lessons and prospects for the development of individualized cancer therapeutics”.<br />
And to start the new year, early in <strong>2009</strong> came the announcement of a new director, Jeff Trent, and a new alliance with the<br />
Translational Genomics Research Institute (TGen); more about that below.<br />
Personnel<br />
Kudos and congratulations go to Craig Webb and Michael Weinreich, who were promoted to Senior <strong>Scientific</strong> Investigator<br />
in September 2008. Craig’s Program of Translational Medicine is developing the infrastructure and biomarkers to bring into<br />
practice individualized medical treatment of diseases like cancer, with the expectation of more-effective treatments from this<br />
approach. Michael’s Laboratory of Chromosome Replication studies molecules that control or regulate the copying of DNA<br />
within a cell and how alterations in the process are related to cancer.<br />
Also in September, Steve Triezenberg, the Dean of the VAI Graduate School and head of the Laboratory of Transcriptional<br />
Regulation, was named Director of the Van Andel Education Institute, succeeding Gordon Van Harn. Our congratulations<br />
to Steve on this new hat to wear. We also congratulate Gordon for his extraordinary contributions in building the Van Andel<br />
Education Institute.<br />
The past year also brought appointments to Brian Haab, who became a member of the Editorial Advisory Board of the<br />
Journal of Proteome Research, and to Bart Williams, who was named to the NIH Skeletal Biology Development and<br />
Disease Study Section.<br />
2
VARI | <strong>2009</strong><br />
We continued to receive grant funding from both federal agencies and other funding organizations in 2008. Brian Haab received<br />
a three-year R33 award from the National Cancer Institute for his project “Defining Secreted Glycan Alterations in Pancreatic<br />
Cancer”. Early in 2008, Cindy Miranti was awarded a three-year DOD grant to study “Mechanisms of KAI1/CD82-Induced<br />
Prostate Cancer Metastasis”. One of her students, Laura Lamb, also received a grant for two years for the project “Survival<br />
Signaling in Prostate Cancer: Role of Androgen Receptor and Integrins in Regulating Survival”.<br />
Brian Cao received 18 months of funding from the Lustgarten Foundation for Pancreatic Cancer Research for his project to<br />
“Generate Monoclonal Antibodies (mAbs) against Pancreatic Cancer Bio-marker Proteins”. Nicholas Duesbery received two<br />
awards, one from the Pardee Foundation for the project “Tumor Endothelial Response to MKK Inhibition”, and another for “Pilot<br />
Investigation of the Causes of Hemangiosarcoma in Clumber Spaniels”. The Vande Woude lab received a grant from the Breast<br />
Cancer Research Foundation for “Met – An Important New Target for Breast Cancer”.<br />
Art Alberts, Brian Cao, and Greg Cavey were recipients of grants from the Michigan Economic Development Corporation during<br />
2008. Bin Teh has been funded to study “Expression Profiling of Renal Cell Carcinoma Utilizing Tissue from CALGB 90206” via<br />
the Roswell Park Cancer Institute. Craig Webb received funds for three years of work to be done on “The Ivy-Genomics-Based<br />
Medicine Project”, via the Translational Genomics Research Institute (TGen).<br />
Chih-Shia Lee of the Duesbery lab and Tingting Yue of the Haab lab received travel awards from the AACR and the Society for<br />
Glycobiology, respectively.<br />
Big Changes<br />
On February 11, <strong>2009</strong>, Van Andel Institute (VAI) announced my retirement from the role of research director. I am proud to<br />
have been a part of VAI’s growth and development over the course of ten wonderful years, watching exciting research unfold<br />
and getting to know the remarkable people and minds of the Van Andel Institute and the Grand Rapids community. We have<br />
built a truly special place, and it is especially gratifying to look at the life sciences construction surrounding the Institute and<br />
know that we have inspired a phenomenon that will benefit patients and families in West Michigan and around the world. With<br />
extraordinary construction to accommodate the Michigan State University College of Human Medicine, the Spectrum Health<br />
and St. Mary’s cancer centers, the expansion of the Helen DeVos Children’s Hospital, and our own expansion, it is truly one of<br />
the most exciting times for Grand Rapids, the state of Michigan, and our nation.<br />
Dr. Jeffrey Trent succeeds me as president and research director while retaining his roles at TGen in Phoenix, Arizona. I have<br />
known Dr. Trent professionally for nearly 20 years. We overlapped at NIH, and I have always admired him as one of the nation’s<br />
leading scientists. This is a very special moment for both institutions and is the right moment and the right place for their perfect<br />
fit to flourish.<br />
I will retain my role as head of the Laboratory of Molecular Oncology at VAI, achieving a long-held desire to return to the lab<br />
full-time. I look forward to being a witness to the Institute’s next phase of growth as we open the Phase II building expansion<br />
and deepen our partnership with TGen.<br />
3
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
4
Van Andel<br />
Research Institute ®<br />
The NEW Alliance for Precision Medicine<br />
About Van Andel<br />
Research Institute<br />
Dr. George<br />
Vande Woude<br />
Director of VARI’s<br />
Laboratory of<br />
Molecular Oncology<br />
On December 1, <strong>2009</strong>,<br />
Van Andel Research Institute<br />
(VARI) and the Translational<br />
Genomics Research Institute<br />
(TGen) will complete a<br />
strategic alliance and affiliation<br />
agreement, enabling both to<br />
maximize their contributions to<br />
science and health. Under the<br />
agreement, Dr. Jeffrey Trent,<br />
TGen’s President and Research<br />
Director, also will become<br />
VARI’s President and Research<br />
Director. Dr. Trent will replace<br />
Dr. George Vande Woude, who<br />
in 1998 was appointed the<br />
founding Director of VARI. Dr.<br />
Vande Woude, a member of the<br />
prestigious National Academy<br />
of Sciences, will remain at VARI<br />
as head of the Laboratory of<br />
Molecular Oncology, allowing<br />
him to achieve a long-held<br />
desire to return to the lab<br />
full-time.<br />
Dr. Jeffrey Trent<br />
President and<br />
Research Director,<br />
VARI and TGen<br />
Established by Jay and Betty Van Andel in 1996,<br />
Van Andel Institute (VAI)<br />
is an independent<br />
research and educational organization<br />
based<br />
in Grand Rapids, Michigan, dedicated to<br />
preserving, enhancing and expanding the<br />
frontiers of medical science, and to achieving<br />
excellence in education by probing fundamental<br />
issues of education and the learning process.<br />
VARI,<br />
the research arm of VAI,<br />
is dedicated to<br />
studying the genetic, cellular and molecular<br />
origins of cancer,<br />
Parkinson’s and other diseases<br />
and working to translate those findings into<br />
effective<br />
therapies. This is accomplished<br />
through the work of over 250 scientists and<br />
staff<br />
in 18 on-site laboratories, in laboratories<br />
in Singapore and Nanjing, and in collaborative<br />
partnerships that span the globe.<br />
For additional information, visit: www.vai.org<br />
About the Translational<br />
Genomics Research<br />
Institute<br />
The Tra<br />
nslational Genomics Research Institute<br />
(TGen) is a non-profit biomedical research<br />
institute based in Phoenix, Arizona, focused<br />
on research that can help patients with<br />
cancer,<br />
neurological disorders, diabetes and<br />
other debilitating conditions. Working with<br />
a worldwide network of collaborators in the<br />
scientific and medical communities, TGen<br />
researchers study the genetic components of<br />
both common and complex diseases. Through<br />
genomic analysis, we learn how DNA, genes<br />
and proteins – the microscopic building block<br />
of life – can affec<br />
t human health. Our 41 lead<br />
investigators and nearly 300 support personnel<br />
at sites in Phoenix, Scottsdale and Flagstaff<br />
,<br />
Arizona, are dedicated to improving patient<br />
care and quality of life through precision<br />
medicine, best defined as the right therapy,<br />
for<br />
the right patient, at the right time.<br />
For additional information,<br />
visit: www.tgen.org
Why the Translational Genomics Research Institute?<br />
Translational Genomics Research Institute<br />
Phoenix, Arizona<br />
TGen presents numerous<br />
opportunities for innovative<br />
scientists and physicians at various<br />
career levels.<br />
Our faculty have access to the latest<br />
technologies – often serving as a test<br />
site for new technology platforms<br />
– for studying genes and proteins,<br />
including advances in next-generation<br />
sequencing techniques as applied to<br />
medical benefit.<br />
In addition to TGen’s research into the<br />
genetic basis of cancer,<br />
neurological<br />
conditions and metabolic disorders,<br />
TGen also plays a role in national<br />
security and bio-defense at TGen<br />
North, our facility in Flagstaff,<br />
Arizona,<br />
under the leadership of internationally<br />
recognized pathogen expert, Dr.<br />
Paul<br />
Keim.<br />
TGen’s Clinical Research Service<br />
(TCRS) at Scottsdale Healthcare<br />
provides TGen with a clinical research<br />
site. Dr. Daniel Von<br />
Hoff,<br />
TGen’s<br />
Physician-in-Chief, also serves as<br />
Chief <strong>Scientific</strong> Officer for TCRS,<br />
where clinicians focus on clinical trials<br />
with targeted agents and genomicsbased<br />
individualized therapy. TCRS,<br />
with an initial focus on cancer,<br />
allows<br />
the unique opportunity for TGen<br />
to transition its laboratory-based<br />
research to patient care centered on<br />
individualized therapy. With nearly<br />
25 active clinical trials for advanced<br />
and/or rare cancers, TCRS is one<br />
of the nation’s leading centers for<br />
PhaseII oncology trials.<br />
TGen’s innovations have resulted in<br />
several partnerships and non-profit<br />
as well as for-profit spin-offs<br />
involving:<br />
physician resources, molecular<br />
profiling, business consulting,<br />
venture capital, drug development<br />
and clinical trials. In partnership with<br />
ASU’s Biodesign Institute, Seattle’s<br />
Fred Hutchinson Cancer Research<br />
Institute and Seattle’s Institute for<br />
Systems Biology,<br />
TGen most recently<br />
established the Partnership for<br />
Personalized Medicine.<br />
The goal of TGen’s education and<br />
outreach programs is to increase<br />
the working knowledge of genomics<br />
within the community at large and to<br />
help educate, train and excite the next<br />
generation of scientists. The Institute’s<br />
roleineducationcontinuouslyevolves,<br />
and currently includes programs<br />
for high school, undergraduate and<br />
graduate students, pre- and postdoctoral<br />
students, and fellowships.<br />
“There are a lot of exceptionally talented scientists at TGen; there are<br />
particular strengths in translational research and in equipment related<br />
to that strength. This includes the infrastructure to perform and<br />
organize clinical trials.’’<br />
— Dr. Bart Williams<br />
Head of VARI’s Laboratory of Cell Signaling and Carcinogenesis, following his visits to TGen<br />
Van Andel<br />
Research Institute ®<br />
333 Bostwick Ave NE<br />
Grand Rapids, MI 49503<br />
(616) 234 5000 | www.vai.org<br />
445 N 5th Street<br />
Phoenix, AZ 85004<br />
(602) 343 8400 | www.tgen.org
VARI | <strong>2009</strong><br />
Laboratory <strong>Report</strong>s<br />
7
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Arthur S. Alberts, Ph.D.<br />
Laboratory of Cell Structure and Signal Integration<br />
In 1993, Dr. Art Alberts received his Ph.D. in Physiology and Pharmacology at the University of<br />
California, San Diego School of Medicine, where he studied with Jim Feramisco. Dr. Alberts trained<br />
as a postdoctoral fellow from 1994 to 1997 with Richard Treisman at the Imperial Cancer Research<br />
Fund in London, England, where Dr. Treisman is the current Director. From 1997 through 1999, Dr.<br />
Alberts was an Assistant Research Biochemist in the laboratory of Frank McCormick at the University<br />
of California, San Francisco. In January 2000, Dr. Alberts joined VARI as a <strong>Scientific</strong> Investigator; he<br />
was promoted in 2006 to Senior <strong>Scientific</strong> Investigator. Also in 2006, he established and became the<br />
Director of the Flow Cytometry core facility.<br />
Staff Students Visiting Scientists<br />
Kathryn Eisenmann, Ph.D.<br />
Leanne Lash-Van Wyhe, Ph.D.<br />
Richard A. West, M.S.<br />
Susan Kitchen, B.S.<br />
Debra Guthrey<br />
Kellie Leali<br />
Aaron DeWard, B.S.<br />
Jonathan Rawson<br />
Albert Rodriguez<br />
Sara Sternberger<br />
Katja Strunk<br />
Stephen Matheson, Ph.D.<br />
Brad Wallar, Ph.D.<br />
8
VARI | <strong>2009</strong><br />
Research Interests<br />
The Laboratory of Cell Structure and Signal Integration is devoted to understanding how defects in cellular architecture affect<br />
the progression to malignancy and support the tumorigenic platform. The driving hypothesis is that the cytoskeleton does not<br />
only structurally support cell morphology, division, and migration, but with its dynamic nature, it organizes intracellular signaling<br />
networks in order to effectively interpret proliferative and migratory responses to extracellular cues. On a molecular basis, we<br />
are interested in how cells build and control the cytoskeletal assembly machines and how these molecular machines work in<br />
concert within the cell. Through combined molecular, cellular, and genetic approaches, the ultimate goal of the lab is identifying<br />
defective nodes in the networks governing cytoskeletal remodeling in order to improve diagnosis and devising molecular tools<br />
to correct the defective circuits.<br />
Our focus is the role of Rho GTPases in signal transduction networks that control cell proliferation and motility. These highly<br />
conserved molecular switches act within growth factor responses by alternating between GTP- and GDP-bound forms. Upon<br />
GTP binding, Rho proteins undergo a conformational change that allows them to bind to and modulate the activity of effectors<br />
that remodel cell shape, drive motility and division, or alter gene expression patterns. One set of GTPase effector proteins acts<br />
as machines that assemble components of the cytoskeleton. The mammalian Diaphanous-related formin (mDia) family of actinnucleating<br />
proteins initiate and control the elongation of new actin filaments. The three conserved mDia proteins (mDia1–3),<br />
along with insect Diaphanous protein and their budding yeast counterpart Bni1p, are canonical members of the formin family.<br />
With our discovery of one of the first formin proteins, mDia2, we have taken a leading role in their characterization.<br />
To study the role of mDia1 in vivo, the murine Drf1 gene was knocked out by conventional gene-targeting methods. Both Drf1 +/–<br />
and Drf1 –/– mice become progressively lympho- and myelodysplastic. Drf1-targeted mice are prone to developing tumors;<br />
cancers observed thus far include various leukemias, monocytosis, and plasmocytomas. Overall, mice lacking one or both<br />
Drf1 alleles phenocopy human myelodysplastic syndrome. Numerous defects in cytoskeletal remodeling have been observed<br />
in immune cells, including impaired T cell adhesion, impaired migration, and the appearance of supernumerary centrosomes,<br />
which are indicative of failed cell division. These results have been published in the Journal of Biological Chemistry, Cancer<br />
Research, and Oncogene.<br />
Overall, the mDia1 knock-out phenotype resembles human chronic myeloproliferative syndrome (MPS) and myelodysplastic<br />
syndrome (MDS). Both MPS and MDS have been characterized as preleukemic states, with variable lymphopenia, excess<br />
or dysfunctional erythrocytes, chronic myelomonocytic leukemia, ineffective hematopoiesis, and, in some cases, advancing<br />
myelofibrosis. Instances of neutrophilic dermatoses (Sweet syndrome) can also accompany MDS and MPS. MDS is a frequent<br />
hematologic disorder that typically affects older patients and is thought to be a stem cell disorder. Dysplastic features of<br />
the nucleus or cytoplasm, as observed in the mDia1 knock-out mice, and altered cellularity of the bone marrow are also<br />
characteristic of MDS. The effect of Drf1 gene targeting and the resulting mDia1 knock-out suggests that the DRF1 gene for<br />
human mDia1 is affected in MPS, MDS, or other preleukemic pathologies. Ongoing studies are focused on examining if defects<br />
in the human gene encoding mDia1 might be defective in MDS patients.<br />
9
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Recent Publications<br />
DeWard, Aaron D., and Arthur S. Alberts. <strong>2009</strong>. Ubiquitin-mediated degradation of the formin mDia2 upon completion of<br />
cell division. Journal of Biological Chemistry 284(30): 20061–20069.<br />
DeWard, Aaron D., Kellie Leali, Richard A. West, George C. Prendergast, and Arthur S. Alberts. <strong>2009</strong>. Loss of RhoB<br />
expression enhances the myelodysplastic phenotype of mammalian Diaphanous-related formin mDia1 knockout mice.<br />
PLoS One 4(9): e7102.<br />
Eisenmann, K.M., K.J. Dykema, S.F. Matheson, N.F. Kent, A.D. DeWard, R.A. West, R. Tibes, K.A. Furge, and A.S. Alberts.<br />
<strong>2009</strong>. 5q– Myelodysplastic syndromes: chromosome 5q genes direct a tumor suppression network sensing actin dynamics.<br />
Oncogene 28(39): 3429–3441.<br />
Shi, Yongquan, Baoxia Dong, Helen Miliotis, Junye Liu, Arthur S. Alberts, Jinyi Zhang, and Katherine A. Siminovitch. <strong>2009</strong>.<br />
Src kinase Hck association with the WASp and mDia1 cytoskeletal regulators promotes chemoattractant-induced Hck<br />
membrane targeting and activation in neutrophils. Biochemistry and Cell Biology 87(1): 207–216.<br />
DeWard, Aaron D., and Arthur S. Alberts. 2008. Microtubule stabilization: formins assert their independence.<br />
Current Biology 18(14): R605–R608.<br />
Kamasani, Uma, James B. DuHadaway, Arthur S. Alberts, and George C. Prendergast. 2007. mDia function is critical for the<br />
cell suicide program triggered by farnesyl transferase inhibition. Cancer Biology & Therapy 6(9): 1422–1427.<br />
From left: Matheson, DeWard, West, Kempston, Kitchen, Alberts, Leali, Rodriguez, Lash-Van Wyhe<br />
10
VARI | <strong>2009</strong><br />
Brian Cao, M.D.<br />
Laboratory of Antibody Technology<br />
Dr. Cao obtained his M.D. from Peking University Medical Center, People’s Republic of China, in 1986.<br />
On receiving a CDC fellowship award, he was a visiting scientist at the National Center for Infectious<br />
Diseases, Centers for Disease Control and Prevention in Colorado (1991–1994). He next served as a<br />
postdoctoral fellow at Harvard (1994–1995) and at Yale (1995–1996). From 1996 to 1999, Dr. Cao was<br />
a Scientist Associate in charge of the Monoclonal Antibody Production Laboratory at the Advanced<br />
BioScience Laboratories–Basic Research Program at the National Cancer Institute, Frederick Cancer<br />
Research and Development Center, Maryland. Dr. Cao joined VARI as a Special Program Investigator<br />
in June 1999 and was promoted to Senior <strong>Scientific</strong> Investigator in July 2006.<br />
Staff Students Visiting Scientists<br />
Quliang Gu, Ph.D.<br />
Ping Zhao, Ph.D.<br />
Tessa Grabinski, B.S.<br />
Amy Nelson<br />
Ximin Chen, M.S.<br />
Guipeng Ding, M.S.<br />
Hong Lin, M.S.<br />
Rui Sun, M.S.<br />
Xiaoting Wang, M.S.<br />
Victoria Hledin<br />
Jessica Karasiewicz<br />
Jin Zhu, Ph.D.<br />
Yunqian Li, M.S.<br />
11
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
Functioning as an antibody production core facility at VARI, our lab develops state-of-the-art services and technology platforms<br />
for monoclonal antibody (mAb) production and characterization. Antibodies are primary tools of biomedical science. In basic<br />
research, the characterization and analysis of almost any molecule involves the production of specific monoclonal or polyclonal<br />
antibodies that react with it. Antibodies are also widely used in clinical diagnostic applications. Further, antibodies are making<br />
rapid inroads into clinical treatment of a variety of diseases, driven by technological evolution from chimeric and humanized to<br />
fully human antibodies.<br />
Our technologies and services include antigen preparation and animal immunization; peptide design and coupling to protein<br />
carriers; immunization with living or fixed cells; conventional antigen/adjuvant preparation; and immunizing a wide range of<br />
antibody-producing models (including mice, rats, rabbits, and transgenic or knock-out mice). Our work also includes the<br />
generation of hybridomas from spleen cells of immunized mice and rats; hybridoma expansion and subcloning; cryopreservation<br />
of hybridomas; mAb isotyping; ELISA screening of hybridoma supernatants; mAb characterization by immunoprecipitation,<br />
immunohistochemistry, immunofluorescence staining, western blot, FACS, and in vitro bioassays; conjugation of mAbs to<br />
enzymes, biotin/streptavidin, or fluorescent reporters; and development of detection kits such as sandwich ELISA. We contract<br />
our services to biotechnology companies, producing and purifying mAbs for their research and for diagnostic kit development.<br />
We have also taken part over the past year in the following research projects.<br />
• A single neutralizing mAb against the HGF/SF alpha domain. Hepatocyte growth factor/scatter factor (HGF/<br />
SF) is a multifunctional heterodimeric polypeptide produced by mesenchymal cells; it is an effector of cells<br />
expressing the Met tyrosine kinase receptor. We previously generated a cocktail of three or four neutralizing<br />
mAbs against HGF/SF that significantly inhibited the HGF-Met signaling pathway in Met-expressing<br />
cells. In a glioblastoma multiforme xenograft model, our cocktail showed potent inhibition of tumor growth.<br />
Amgen and others have reported a single anti-HGF/SF b-subunit mAb that is able to inhibit biological activities<br />
of HGF/SF; it is in early clinical trials. We hypothesized that two mAbs that react with different subunits<br />
(a and b) of HGF/SF in combination would have stronger anti-tumor activity than any single antibody. Using<br />
a unique immunization protocol, we have generated a mAb against the HGF/SF a subunit (designated HGF8)<br />
that has neutralizing activity. Our current results show that HGF8 is able to block HGF/SF-induced scattering<br />
of MDCK cells, and in collaboration with the Vande Woude lab, we have shown that HGF8 also significantly<br />
inhibits the Met-HGF/SF signaling pathway in vitro using uPA and cell proliferation assays. The in vivo antitumor<br />
activity of HGF8 is now under investigation in a brain tumor xenograft model using HGF/SF transgenic<br />
mice established by the Vande Woude lab.<br />
• Development of highly specific anti-Met mouse mAbs with potential application for clinical immunohistochemical<br />
diagnosis. In collaboration with Beatrice Knudsen’s lab at the Fred Hutchinson Cancer Research Center,<br />
we have developed a monoclonal antibody, designated MET4, with the goal of accurately and reproducibly<br />
measuring MET in formalin-fixed paraffin-embedded (FFPE) tissues. MET4 was selected as the best probe<br />
from a pool of MET-avid monoclonal antibodies, based on its specific staining pattern in FFPE preparations of<br />
normal human prostate tissues. The reliability of MET4 immunohistochemistry was assessed by comparing<br />
MET4-IHC in FFPE cell pellets with immunoblotting analysis, which demonstrated a high avidity of MET4 for<br />
formalin-treated MET. These properties encourage further development of MET4 as a multipurpose molecular<br />
diagnostic reagent to help guide the selection of individual patients being considered for treatment with MET<br />
antagonistic drugs.<br />
12
VARI | <strong>2009</strong><br />
• Generation of monoclonal antibodies against pancreatic cancer biomarkers. In March 2008, the Lustgarten<br />
Foundation officially launched the Pancreatic Cancer Biomarker Development Initiative. Identifying key pancreatic<br />
cancer biomarkers and producing antibodies against them is the first step toward developing a blood<br />
test for this disease. A consortium of investigators representing four leading cancer research organizations—<br />
including the Canary Foundation, Dana-Farber Cancer Institute, University of California, San Francisco, and<br />
Van Andel Research Institute—will study a total of 60 candidate biomarkers. We have been assigned 15<br />
biomarkers and funding for 18 months. The project is to develop monoclonal antibodies against those biomarkers,<br />
including paired mAbs for sandwich ELISA development, mAbs specifically for western blotting and<br />
immunohistochemical study, etc. All biomarkers need to be expressed and purified by the lab. This project<br />
also requires collaboration with other labs and core facilities. For example, we will collaborate with Brian<br />
Haab’s VARI lab to identify paired mAbs for sandwich ELISA development using antibody array technology.<br />
We will also use James Resau’s VARI histology/pathology core and tissue microarray technology to characterize<br />
the mAbs that work best for immunohistochemical staining.<br />
Recent Publications<br />
Knudsen, Beatrice S., Ping Zhao, James Resau, Sandra Cottingham, Ermanno Gherardi, Eric Xu, Bree Berghuis,<br />
Jennifer Daugherty, Tessa Grabinski, Jose Toro, et al. <strong>2009</strong>. A novel multipurpose monoclonal antibody for evaluating human<br />
c-Met expression in preclinical and clinical settings. Applied Immunohistochemistry and Molecular Morphology 17(1): 56–67.<br />
Nguyen, Melissa L., Sherry R. Crowe, Sridevi Kurella, Simon Teryzan, Brian Cao, Jimmy D. Ballard, Judith A. James, and<br />
A. Darise Farris. <strong>2009</strong>. Sequential B cell epitopes of Bacillus anthracis lethal factor bind lethal toxin–neutralizing antibodies.<br />
Infection and Immunity 77(1): 162–169.<br />
Wang, Xin, and Brian B. Cao. <strong>2009</strong>. Screening of specific internalization Fab fragments from human naïve phage library<br />
by combinational bio-panning. In Therapeutic Antibodies: Methods and Protocols, Antony S. Dimitrov, ed. Methods in<br />
Molecular Biology series, Vol. 525. New York: Humana Press, pp. 161–174.<br />
Chen, Jindong, Kunihiko Futami, David Petillo, Jun Peng, PengFei Wang, Jared Knol, Yan Li, Sok Kean Khoo, Dan Huang,<br />
Chao-Nan Qian, et al. 2008. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia.<br />
PLoS One 3(10): e3581.<br />
Xie, Qian, Ryan Thompson, Kim Hardy, Lisa DeCamp, Bree Berghuis, Robert Sigler, Beatrice Knudsen, Sandra Cottingham,<br />
Ping Zhao, Karl Dykema, et al. 2008. A highly invasive human glioblastoma pre-clinical model for testing therapeutics.<br />
Journal of Translational Medicine 6: 77.<br />
From left: Ding, Lin, Wang, Cao, Grabinski, Zhu, Nelson, Zhao<br />
13
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Gregory S. Cavey, B.S.<br />
Laboratory of Mass Spectrometry and Proteomics<br />
Mr. Cavey received his B.S. degree from Michigan State University in 1990. Prior to joining VARI he was<br />
employed at Pharmacia in Kalamazoo, Michigan, for nearly 15 years. As a member of a biotechnology<br />
development unit, he was group leader for a protein characterization core laboratory. More recently<br />
as a research scientist, he was principal in the establishment and application of a state-of-the-art<br />
proteomics laboratory for drug discovery. Mr. Cavey joined VARI as a Special Program Investigator in<br />
July 2002.<br />
Staff<br />
Paula Davidson, M.S.<br />
Caryn Lehner, M.S.<br />
Matthew Welsh, B.S.<br />
Debra Guthrey<br />
14
VARI | <strong>2009</strong><br />
Research Interests<br />
Mass spectrometry–based proteomics is now an important and widespread tool in basic and clinical research. In 2005, VARI<br />
purchased a Waters quadrupole time-of-flight (Q-Tof) mass spectrometry system that remains at the cutting edge of many<br />
research applications. This equipment allows us to provide routine mass spectrometry services and to develop new services<br />
such as protein profiling for biomarker discovery and protein phosphorylation analysis.<br />
Protein identification and protein molecular weight determination are routine services performed on sub-microgram amounts<br />
of material to address a wide variety of biological questions. Protein identification via mass spectrometry is mainly used to<br />
identify novel protein-protein interactions and can be performed on proteins in SDS-PAGE gels or in solutions. Molecular<br />
weight determination of protein solutions is typically used to confirm the expression and purification of recombinant proteins to<br />
be used as reagents in x-ray crystallographic experiments or drug screening/cell-based assays. Our research emphasis is on<br />
1) developing liquid chromatography–mass spectrometry (LC-MS) protein profiling analysis for systems biology research and<br />
biomarker discovery and 2) improving methods for identifying and quantifying the phosphorylation of proteins.<br />
LC-MS protein profiling<br />
Our lab collaborates with Waters Corporation, a major manufacturer of mass spectrometry and HPLC equipment, to evaluate<br />
and improve existing methods while applying LC-MS to the research efforts of VARI scientists and of external clients. Our<br />
LC-MS system employs a novel data acquisition method unique to Waters mass spectrometers, termed LC-MS E , whereby<br />
quantitative and qualitative data are collected in a single analysis. Protein samples are first digested into peptides using<br />
trypsin and then analyzed by reverse-phase nanoscale LC-MS. Recording peptide mass, HPLC retention time, and intensity<br />
as measured in the mass spectrometer, we digitize the data to allow comparisons across samples. Quantitation is based on<br />
measuring and comparing the chromatographic peak area for each peptide across samples. Qualitative protein identification<br />
data is collected in a multiplexed, non-intensity-biased fashion concurrent with quantitative data. One current pilot project is a<br />
time-course analysis of protein secretion (secretome) from mouse 3T3-L1 pre-adipocytes as they differentiate in response to<br />
treatment with dexamethasone/insulin, versus the response to the PPARg antagonist rosiglitazone. A second study is of the<br />
secretome of a cell line model of cachexia.<br />
In addition to mechanism-of-action studies, our goal is to use LC-MS to discover candidate biomarkers of disease. Current<br />
research efforts focus on sample processing techniques to reproducibly fractionate highly complex samples such as blood<br />
plasma, tissue, and urine to allow quantitative analysis. Replicate LC-MS analysis of carefully chosen samples and multivariate<br />
data analysis will allow us to differentiate between normal biological variation and disease.<br />
Protein phosphorylation analysis<br />
Mapping post-translational protein modifications such as phosphorylation is an important yet difficult undertaking. In cancer<br />
research, phosphorylation regulates many protein pathways that could serve as targets for drug therapy. In recent years,<br />
mass spectrometry has emerged as a primary tool in determining site-specific phosphorylation and relative quantitation.<br />
Phosphorylation analysis is complicated by many factors, but principally by the low-stoichiometry modifications that may<br />
regulate pathways: we are sometimes dealing with 0.01% or less of phosphorylated protein among a large excess of a<br />
nonphosphorylated counterpart.<br />
15
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
As with most mass spectrometry–based methods, the mapping of phosphorylation sites on proteins begins by enzymatically<br />
digesting protein into peptides using trypsin, Lys-C, Staph V8, or chymotrypsin. Peptides are separated by nanoscale reversephase<br />
HPLC and analyzed by on-line electrospray ionization on a Q-Tof mass spectrometer. Samples are analyzed using MS E<br />
data acquisition. MS E toggles the collision energy in the mass spectrometer between high and low every second throughout<br />
the analytic run. Low-collision-energy data acquisition allows peptide mass to be recorded at high sensitivity with high mass<br />
accuracy to implicate phosphorylation based on mass alone. The peptide intensity measured in the mass spectrometer is<br />
also recorded and used for relative quantitation in time course studies. During high-collision-energy acquisition, all peptides<br />
are fragmented to identify the protein(s) that the peptides were liberated from and to locate specific phosphorylated amino<br />
acids. MS E differs from other mass spectrometry approaches because fragmentation occurs for all peptides, not just for the<br />
most abundant peptides. We recently used this method for mapping phosphorylation sites on RhoA and RhoC following in<br />
vitro phosphorylation by protein kinase C epsilon (PKCe). We are currently analyzing RhoA and RhoC in a head and neck<br />
squamous cell carcinoma tissue culture model with or without the expression of PKCe using siRNA knock-down.<br />
External Collaborators<br />
Gary Gibson, Henry Ford Hospital, Detroit, Michigan<br />
Quintin Pan, Ohio State University Comprehensive Cancer Center<br />
Waters Corporation<br />
Recent Publications<br />
Yang, Maozhou, Xinli Wang, Liang Zhang, Chiyang Yu, Bingbing Zhang, William Cole, Greg Cavey, Paula Davidson, and<br />
Gary Gibson. 2008. Demonstration of the interaction of transforming growth factor beta 2 and type X collagen using a<br />
modified tandem affinity purification tag. Journal of Chromatography B 875(2): 493–501.<br />
From left to right, standing: Cavey, Lehner, Davidson; seated: Guthrey, Welsh<br />
16
VARI | <strong>2009</strong><br />
Nicholas S. Duesbery, Ph.D.<br />
Laboratory of Cancer and Developmental Cell Biology<br />
Nick Duesbery received a B.Sc. (Hon.) in biology (1987) from Queen’s University, Canada, and both his<br />
M.Sc. (1990) and Ph.D. (1996) degrees in zoology from the University of Toronto, Canada, under the<br />
supervision of Yoshio Masui. Before his appointment as a <strong>Scientific</strong> Investigator at VARI in April 1999,<br />
he was a postdoctoral fellow in the laboratory of George Vande Woude in the Molecular Oncology<br />
Section of the Advanced BioScience Laboratories–Basic Research Program at the National Cancer<br />
Institute, Frederick Cancer Research and Development Center, Maryland. Dr. Duesbery was promoted<br />
to Senior <strong>Scientific</strong> Investigator and appointed Deputy Director for Research Operations in 2006.<br />
Staff Students Visiting Scientist<br />
Jennifer Bromberg-White, Ph.D.<br />
Jaclyn Lynem, B.S.<br />
Elissa Boguslawski<br />
Laura Holman<br />
Chih-Shia Lee, M.S.<br />
Danielle Hawkins, B.S.<br />
Emily Olenzek, B.S.<br />
Michelle Dawes<br />
Shannon Moran<br />
Roe Froman, D.V.M.<br />
17
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
Many malignant sarcomas such as fibrosarcomas are refractory to available treatments. However, sarcomas possess unique<br />
vascular properties which indicate they may be more responsive to therapeutic agents that target endothelial function. Mitogenactivated<br />
protein kinase kinases (MKKs) play an essential role in the growth of carcinomas, and we hypothesize that signaling<br />
through multiple MKK pathways is also essential for sarcomas. One objective of our research is to define the role of MKK<br />
signaling in the growth and vascularization of human sarcomas and to determine whether MKK inhibitors can form the basis of<br />
a novel and innovative approach to the treatment of human sarcoma.<br />
In 2008, we published a study showing that inhibition of MKK signaling by lethal toxin (LeTx) caused a rapid and dramatic<br />
decrease in tumor perfusion that was followed by a long-term reduction in tumor vascularization. Follow-up histologic analysis<br />
in collaboration with VARI’s James Resau (Laboratory of Analytical, Cellular, and Molecular Microscopy) showed this acute<br />
decrease in tumor perfusion was caused by increased leakiness of tumor blood vessels. This was unexpected, because antiangiogenic<br />
agents typically lead to a regression of neovascularization over the course of weeks, not hours. Moreover, these<br />
agents typically normalize tumor-associated blood vessels, rendering them less leaky. The results of our study show that while<br />
MKK activity is required for tumor cell proliferation, it also plays an important role in tumor vascular function.<br />
With funding from the Elsa Pardee Foundation, we have continued our investigation of the effects of MKK inhibition on vascular<br />
function in sarcomas. In parallel, Jenn Bromberg-White, a postdoctoral fellow, has begun an investigation into the roles MKK<br />
pathways play in the formation of vascular networks in the developing mouse eye, while Chih-Shia Lee, a graduate student, is<br />
performing a detailed study of the individual contributions of MKK pathways to melanoma survival.<br />
In 2008 we also began a new project on hemangiosarcomas, a soft-tissue tumor for which there are currently no effective<br />
treatments. Although rare in humans, hemangiosarcomas are relatively common in certain breeds of dogs such as Golden<br />
Retrievers, German Shepherds, and Clumber Spaniels. Hemangiosarcomas seem to run in families, indicating that there is an<br />
underlying hereditary or genetic component to this disease.<br />
To study these tumors, we have established the Canine Hereditary Cancer Consortium (CHCC). With the support of the<br />
American Kennel Club Canine Health Foundation (AKC CHF Grant 1114) and the Clumber Spaniel Health Foundation, the<br />
CHCC will take advantage of new genetic resources and technologies at Van Andel Research Institute to develop genetic<br />
screens, diagnostic tests, and treatments for hereditary canine cancers, as well as to gain insight into the biology of human<br />
disease. In our pilot proposal, we have focused on hemangiosarcomas in Clumber Spaniels; later we will include other<br />
breeds and additional hereditary cancers. We will analyze collected DNA and RNA samples from Clumber Spaniels for genetic<br />
patterns that are associated with this disease. These patterns may form the basis of genetic tests that can tell us whether a<br />
particular dog is a carrier of a defective gene that will cause cancer. Also, these studies may provide important clues about<br />
hemangiosarcomas in humans. Key laboratories participating in this project include the Laboratory of Cancer Genetics; the<br />
Laboratory of Analytical, Cellular, and Molecular Microscopy; the Laboratory of Computational Biology; and the Laboratory of<br />
Cancer & Developmental Cell Biology. Dr. Roe Froman, D.V.M., is our consulting veterinarian.<br />
18
VARI | <strong>2009</strong><br />
Recent Publications<br />
Alfano, Randall W., Stephen H. Leppla, Shihui Liu, Thomas H. Bugge, Cynthia J. Meininger, Terry C. Lairmore,<br />
Arlynn F. Mulne, Samuel H. Davis, Nicholas S. Duesbery, and Arthur E. Frankel. <strong>2009</strong>. Matrix metalloproteinase–<br />
activated anthrax lethal toxin inhibits endothelial invasion and neovasculature formation during in vitro morphogenesis.<br />
Molecular Cancer Research 7(4): 452–461.<br />
Bromberg-White, Jennifer L., Elissa Boguslawski, and Nicholas S. Duesbery. <strong>2009</strong>. Perturbation of mouse retinal vascular<br />
morphogenesis by anthrax lethal toxin. PLoS One 4(9): e6956.<br />
Alfano, Randall W., Stephen H. Leppla, Shihui Liu, Thomas H. Bugge, Meenhard Herlyn, Keiran S. Smalley,<br />
Jennifer L. Bromberg-White, Nicholas S. Duesbery, and Arthur E. Frankel. 2008. Cytotoxicity of the matrix metalloproteinase–<br />
activated anthrax lethal toxin is dependent on gelatinase expression and B-RAF status in human melanoma cells.<br />
Molecular Cancer Therapeutics 7(5): 1218–1226.<br />
Ding, Yan, Elissa A. Boguslawski, Bree D. Berghuis, John J. Young, Zhongfa Zhang, Kim Hardy, Kyle Furge, Eric Kort,<br />
Arthur E. Frankel, Rick V. Hay, et al. 2008. Mitogen-activated protein kinase kinase signaling promotes growth and<br />
vascularization of fibrosarcoma. Molecular Cancer Therapeutics 7(3): 648–658.<br />
Kuo, Shu-Ru, Mark C. Willingham, Sarah H. Bour, Elissa A. Andreas, Seong Kyu Park, Carney Jackson, Nicholas S. Duesbery,<br />
Stephen H. Leppla, Wei-Jen Tang, and Arthur E. Frankel. 2008. Anthrax toxin–induced shock in rats is associated with<br />
pulmonary edema and hemorrhage. Microbial Pathogenesis 44(6): 467–472.<br />
From left: Lee, Boguslawski, Holman, Duesbery, Froman, Bromberg-White;<br />
foreground: D Too, a Clumber Spaniel<br />
19
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Bryn Eagleson, B.S., RLATG<br />
Vivarium and Transgenics Program<br />
Bryn Eagleson began her career in laboratory animal services in 1981 with Litton Bionetics at the<br />
National Cancer Institute’s Frederick Cancer Research and Development Center (NCI–FCRDC) in<br />
Maryland. In 1983, she joined the Johnson & Johnson Biotechnology Center in San Diego, California. In<br />
1988, she returned to NCI–FCRDC, where she continued to develop her skills in transgenic technology<br />
and managed the transgenic mouse colony. In 1999, she joined VARI as the Vivarium Director and<br />
Transgenics Special Program Manager.<br />
Technical Staff Animal Caretaker Staff IACUC Coordinator<br />
Lisa DeCamp, B.S.<br />
Dawna Dylewski, B.S.<br />
Audra Guikema, B.S., L.V.T.<br />
Tristan Kempston, B.S.<br />
Angie Rogers, B.S.<br />
Elissa Boguslawski, RALAT<br />
Tina Schumaker, ALAT<br />
20<br />
Sylvia Marinelli, Vivarium Supervisor<br />
Crystal Brady<br />
Neil Brandow<br />
Jarred Grams<br />
Rishard Moody<br />
Janelle Post<br />
Drew Rapp<br />
Bobbie Vitt<br />
Alma Klotz
VARI | <strong>2009</strong><br />
Research Interests<br />
The goal of the vivarium and the transgenics program is to develop, provide, and support high-quality mouse modeling services<br />
for the Van Andel Research Institute investigators, Michigan collaborators, and the greater research community. We use three<br />
Topaz Technologies software products—Granite, Scion, and Topaz Protocols and Reviews—for integrated management of the<br />
vivarium finances, the mouse breeding colony, and the Institutional Animal Care and Use Committee (IACUC) protocols and<br />
records, respectively. Imaging equipment, such as the PIXImus mouse densitometer and the ACUSON Sequoia 512 ultrasound<br />
machine, is available for noninvasive imaging of mice. Also provided by the vivarium technical staff are an extensive xenograft<br />
model development and analysis service, rederivation, surgery, dissection, necropsy, breeding, and health-status monitoring.<br />
Transgenics<br />
Fertilized eggs contain two pronuclei, one that is derived from the egg and contains the maternal genetic material and one<br />
derived from the sperm that contains the paternal genetic material. As development proceeds, these two pronuclei fuse,<br />
the genetic material mixes, and the cell proceeds to divide and develop into an embryo. Transgenic mice are produced by<br />
injecting small quantities of foreign DNA (the transgene) into a pronucleus of a one-cell fertilized egg. DNA microinjected into a<br />
pronucleus randomly integrates into the mouse genome and will theoretically be present in every cell of the resulting organism.<br />
Expression of the transgene is controlled by elements called promoters that are genetically engineered into the transgenic<br />
DNA. Depending on the selection of the promoter, the transgene can be expressed in every cell of the mouse or in specific cell<br />
populations such as neurons, skin cells, or blood cells. Temporal expression of the transgene during development can also<br />
be controlled by genetic engineering. These transgenic mice are excellent models for studying the expression and function of<br />
the transgene in vivo.<br />
Recent Publications<br />
Monks, Douglas Ashley, Jamie A. Johansen, Kaiguo Mo, Pengcheng Rao, Bryn Eagleson, Zhigang Yu, Andrew P. Lieberman,<br />
S. Marc Breedlove, and Cynthia L. Jordan. 2007. Overexpression of wild-type androgen receptor in muscle recapitulates<br />
polyglutamine disease. Proceedings of the National Academy of Sciences U.S.A. 104(46): 18259–18264.<br />
From left: Vitt, Brandow, Marinelli, Brady, Eagleson, Moody, Klotz, Schumaker, Rapp, Boguslawski, Kempston, Dylewski, DeCamp,<br />
Guikema, Grams, Post<br />
21
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Mouse liver cells<br />
Although it looks like a child’s fingerpainting, this is a micrograph of mouse liver cells. Green stain marks endothelial cells, red stain marks the<br />
actin cytoskeleton of fibroblasts, and blue stain marks cell nuclei. Photo by Veronique Schulz of the Miranti lab.<br />
22
VARI | <strong>2009</strong><br />
Kyle A. Furge, Ph.D.<br />
Laboratory of Computational Biology<br />
Dr. Furge received his Ph.D. in biochemistry from the Vanderbilt University School of Medicine in 2000.<br />
Prior to obtaining his degree, he worked as a software engineer at YSI, Inc., where he wrote operating<br />
systems for remote environmental sensors. Dr. Furge did his postdoctoral work in the laboratory of<br />
George Vande Woude. He became a Bioinformatics Scientist at VARI in June of 2001 and a <strong>Scientific</strong><br />
Investigator in May of 2005.<br />
Staff<br />
Karl Dykema, B.A.<br />
Amy Nelson<br />
Students<br />
Craig Johnson, P.S.M.<br />
Jeff Klomp, M.S.<br />
Theresa Gipson<br />
23
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
High-throughput technologies—such as DNA sequencing, gene and protein expression profiling, DNA copy number analysis,<br />
and single nucleotide polymorphism genotyping—produce large amounts of data and have created a need for new tools that<br />
can assist in extracting the significant biological information from these data sets. Bioinformatics and computational biology<br />
are new disciplines that develop methods for the storage, distribution, integration, and analysis of these large data sets. The<br />
Computational Biology laboratory at VARI uses mathematical and computer science approaches to analyze and integrate<br />
complex data sets with a goal of understanding how cancer cells differ from normal cells at the molecular level. In addition,<br />
members of the lab provide assistance in data analysis and other computational projects on a collaborative and/or fee-forservice<br />
basis.<br />
In the past year, the laboratory has taken part in many collaborative projects to further the research efforts at VARI. We have<br />
contributed gene expression analysis to projects ranging from identifying mechanisms of oncogene transformation to identifying<br />
genes associated with drug resistance. In recent work led by the Laboratory of Chromosome Replication, we examined how<br />
the deregulation of genes involved in chromosome replication are associated with the development and progression of several<br />
types of cancer. We have worked closely with the Laboratory of Cancer Genetics in developing gene expression–based models<br />
for the diagnosis and prognosis of renal cell carcinoma. We are also part of a multi-lab project spearheaded by the Laboratory<br />
of Cancer and Developmental Cell Biology to identify and characterize genes associated with the development of hereditary<br />
hemangiosarcomas in canines. Our role in this project focuses on the integration of data from single nucleotide polymorphism,<br />
gene expression, and pathway modeling studies.<br />
In addition to collaborative work, the lab has a particular interest in developing and applying computational models that use<br />
gene expression data to identify large chromosomal abnormalities in cancer cells. In humans, each cell contains a set of<br />
approximately 6 billion DNA bases that are packaged into 46 chromosomes. From these chromosomes, at least 20,000<br />
different types of messenger RNAs (mRNAs) and hundreds of non-coding RNAs (ncRNAs) are produced. Structural changes<br />
in chromosomes, such as translocations, deletions, rearrangements, and amplifications, commonly occur in cancer cells and<br />
likely contribute to the development and progression of the disease through disruptions in RNA production. We are building<br />
computational tools that use RNA expression to both identify chromosomal abnormalities and identify which single RNA (or<br />
set of RNAs), when deregulated, contributes to tumor development. In recent work, these RNA-based models predicted that<br />
high-grade papillary renal cell carcinoma contained a chromosome 8q amplification associated with overexpression of the<br />
c-MYC gene and activation of the MYC transcriptional program. This prediction was subsequently confirmed using molecular<br />
and cell biology experiments, highlighting the potential of gene expression profiling data for building integrative computational<br />
models of tumor development and progression.<br />
The use of RNA-based models has the potential to identify even more-subtle chromosomal changes, such as changes in<br />
chromosome conformation. Examination of gene expression data derived from a subtype of renal cancer, renal oncocytoma,<br />
revealed that the population of RNAs produced from chromosome 19 was significantly up-regulated relative to the RNAs<br />
produced in normal kidney cells. Although no structural abnormality on chromosome 19 was identified, a more detailed<br />
cytogenetic analysis of renal oncocytoma cells showed that the chromosome 19 homologues had become intertwined or<br />
“paired”. The pairing was associated with the changes in the amount of mRNA produced from this chromosome. We are<br />
currently working to determine if chromosome pairing is present in other types of tumor cells and to determine the role of the<br />
chromosomal state in tumor development and progression.<br />
24
VARI | <strong>2009</strong><br />
Recent Publications<br />
Eisenmann, K.M., K.J. Dykema, S.F. Matheson, N.F. Kent, A.D. DeWard, R.A. West, R. Tibes, K.A. Furge, and A.S. Alberts. <strong>2009</strong>.<br />
5q– Myelodysplastic syndromes: chromosome 5q genes direct a tumor suppression network sensing actin dynamics.<br />
Oncogene 28(39): 3429–3441.<br />
Hui, Zhouguang, Maria Tretiakova, Zhongfa Zhang, Yan Li, Xiaozhen Wang, Julie Xiaohong Zhu, Yuanhong Gao, Weiyuan Mai,<br />
Kyle Furge, Chao-Nan Qian, et al. <strong>2009</strong>. Radiosensitization by inhibiting STAT1 in renal cell carcinoma. International Journal<br />
of Radiation Oncology Biology Physics 73(1): 288–295.<br />
Wang, Y., O. Roche, M.S. Yan, G. Finak, A.J. Evans, J.L. Metcalf, B.E. Hast, S.C. Hanna, B. Wondergem, K.A. Furge, et al.<br />
<strong>2009</strong>. Regulation of endocytosis via the oxygen-sensing pathway. Nature Medicine 15(3): 319–324.<br />
Bonte, Dorine, Charlotta Lindvall, Hongyu Liu, Karl Dykema, Kyle Furge, and Michael Weinreich. 2008. Cdc7-Dbf4 kinase<br />
overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia 10(9): 920–931.<br />
Camparo, Philippe, Viorel Vasiliu, Vincent Molinié, Jerome Couturier, Karl J. Dykema, David Petillo, Kyle A. Furge, Eva M.<br />
Comperat, Marick Laé, Raymonde Bouvier, et al. 2008. Renal translocation carcinomas: clinicopathologic, immunohistochemical,<br />
and gene expression profiling analysis of 31 cases with a review of the literature. American Journal of Surgical Pathology<br />
32(5): 656–670.<br />
Chen, Jindong, Kunihiko Futami, David Petillo, Jun Peng, PengFei Wang, Jared Knol, Yan Li, Sok Kean Khoo, Dan Huang,<br />
Chao-Nan Qian, et al. 2008. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia.<br />
PLoS One 3(10): e3581.<br />
Koeman, Julie M., Ryan C. Russell, Min-Han Tan, David Petillo, Michael Westphal, Katherine Koelzer, Julie L. Metcalf,<br />
Zhongfa Zhang, Daisuke Matsuda, Karl J. Dykema, et al. 2008. Somatic pairing of chromosome 19 in renal oncocytoma is<br />
associated with deregulated EGLN2-mediated oxygen-sensing response. PLoS Genetics 4(9): e1000176.<br />
Kort, Eric J., Leslie Farber, Maria Tretiakova, David Petillo, Kyle A. Furge, Ximing J. Yang, Albert Cornelius, and Bin T. Teh. 2008.<br />
The E2F3–Oncomir-1 axis is activated in Wilms’ tumor. Cancer Research 68(11): 4034–4038.<br />
Zhang, Zhong-Fa, Daisuke Matsuda, Sok Kean Khoo, Kristen Buzzitta, Elizabeth Block, David Petillo, Stéphane Richard,<br />
John Anema, Kyle A. Furge, and Bin T. Teh. 2008. A comparison study reveals important features of agreement and disagreement<br />
between summarized DNA and RNA data obtained from renal cell carcinoma. Mutation Research 657(1): 77–83.<br />
From left: Dykema, Furge, Klomp, Nelson<br />
25
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Brian B. Haab, Ph.D.<br />
Laboratory of Cancer Immunodiagnostics<br />
Dr. Haab obtained his Ph.D. in chemistry from the University of California at Berkeley in 1998. He then<br />
served as a postdoctoral fellow in the laboratory of Patrick Brown in the Department of Biochemistry<br />
at Stanford University. Dr. Haab joined VARI as a Special Program Investigator in May 2000, became a<br />
<strong>Scientific</strong> Investigator in 2004, and was promoted to Senior <strong>Scientific</strong> Investigator in 2007.<br />
Staff Students Visiting Scientist<br />
John Buchweitz, Ph.D.<br />
Yi-Mi Wu, Ph.D.<br />
Derek Bergsma, B.S.<br />
Steven Kluck, B.S.<br />
Andrew Porter, B.S.<br />
Amy Nelson<br />
Rob Antecki, B.S.<br />
Kim Babins, B.S.<br />
Carrie Fiebig, B.S.<br />
Lee Heeringa, B.S.<br />
Kevin Maupin, B.A.<br />
Arkadeep Sinha, B.S.<br />
Dan Hekman<br />
Christopher Madziar<br />
Randi VanOcker<br />
Tingting Yue, B.S.<br />
David Nowack, Ph.D.<br />
26
VARI | <strong>2009</strong><br />
Research Interests<br />
All cells secrete molecules that are used to send signals and perform functions in the local and distant spaces of the body. The<br />
molecular secretions of cancer cells often are significantly different from those of their normal counterparts. Our lab studies<br />
particular proteins and carbohydrates secreted by cancer cells in order to understand their roles in cancer progression, as well<br />
as to develop novel clinical tests for the detection and diagnosis of cancer.<br />
Glycoprotein biomarkers for pancreatic cancer<br />
A great need exists for better tools to detect and diagnose incipient pancreatic cancer. Our laboratory is addressing this problem<br />
by taking advantage of a frequently observed molecular feature of pancreatic cancer, i.e., alterations to the carbohydrate side<br />
chains of cell-surface and secreted proteins. Most secreted proteins have carbohydrates known as glycans attached to them,<br />
and some of the secreted proteins with altered carbohydrates are released into the blood of cancer patients. The measurement<br />
of certain secreted glycoproteins, along with their attached glycans, could form the basis of effective diagnostic markers.<br />
A particularly valuable platform for probing glycan variants on specific proteins is the antibody-lectin sandwich array (ALSA),<br />
developed earlier in our laboratory. The method starts with a microarray of antibodies that target various glycoproteins of<br />
interest. A complex biological sample is incubated on the array, resulting in the capture of glycoproteins by the antibodies.<br />
Then the array is probed with a lectin (a protein with carbohydrate-binding activity), which binds to the captured glycoproteins<br />
that bear the lectin’s glycan target. The amount of lectin binding at each antibody indicates the amount of glycan on the<br />
proteins captured by that antibody. Diverse lectins can be used to probe a variety of glycans on a given sample. In addition,<br />
the captured proteins can be probed with antibodies targeting the core proteins, as in a “sandwich” immunoassay, to obtain<br />
the levels of the proteins in parallel assays.<br />
Relative to other technologies, the platform offers a unique combination of capabilities such as reproducible glycan measurements<br />
on specific proteins, high-throughput sample processing, and high-sensitivity detection directly from biological samples.<br />
These features make the platform ideal for glycoprotein-based biomarker studies. A product based on this technology is now<br />
available from GenTel Biosciences (Madison, Wisconsin).<br />
Using this tool, we can now explore the hypotheses that particular glycan structures on specific proteins are found uniquely in<br />
certain disease states and that their measurement yields effective detection of cancer. We have characterized the prevalence<br />
in pancreatic cancer patients of a variety of glycan structures on several types of mucin proteins. Some glycan alterations<br />
were found in a high percentage of the cancer patients but not at all in healthy<br />
Figure 1<br />
subjects. Furthermore, the glycan levels were altered independently of<br />
changes to the protein level, so that measuring both the glycan and protein<br />
level gives improved biomarker performance relative to measuring only protein<br />
levels as in standard immunoassays. The performance of these initial studies<br />
already suggests improvement upon the best current biomarkers for pancreatic<br />
cancer. Now we are working to characterize and develop detection<br />
methods for both the protein forms that carry cancer-associated glycans and<br />
the glycans themselves.<br />
Figure 1. Protein and glycan detection using antibody arrays. a) Array-based<br />
sandwich assays for protein detection. Multiple antibodies are immobilized on a planar<br />
support, and the captured proteins are probed using biotinylated detection antibodies,<br />
followed by fluorescence detection using phycoerythrin-labeled streptavidin.<br />
b) Antibody-lectin sandwich arrays (ALSA). This format is similar to a), but the detection<br />
reagents target the glycans on the capture proteins rather than the core proteins. The<br />
glycans on the immobilized antibodies are chemically derivatized to prevent lectin<br />
binding to those glycans. c) Example antibody array results for core protein detection<br />
(left) and glycan measurement (right). SA-PE, streptavidin-phycoerythrin.<br />
27
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
We work with clinical collaborators at several institutions to address various clinical needs. One such need is to help doctors<br />
make a more accurate diagnosis of patients with suspected pancreatic cancer. Since pancreatic cancer can be difficult to<br />
distinguish from benign conditions of the gastrointestinal tract, highly accurate biomarkers are needed to match patients to the<br />
appropriate procedures at the earliest possible time. We also are developing biomarkers to screen for clinically undetectable<br />
pancreatic cancer. Among populations at an increased risk for developing pancreatic cancer—including those suffering from<br />
chronic pancreatitis or with a family history of pancreatic cancer—an accurate screening test could detect new cancers early<br />
enough to allow more effective treatment. Further, we are testing our novel biomarkers for use in drug trials. Biomarkers that<br />
give early indications of the effectiveness of a candidate drug could accelerate drug trials and better match patients with the<br />
drugs that benefit them most.<br />
Another novel class of biomarker we are developing is for the diagnosis of patients with pancreatic cysts. Cystic lesions of the<br />
pancreas are increasingly being recognized due to the widespread use of high-resolution abdominal imaging. Since certain<br />
cyst types are precursors of invasive cancer, this situation presents an opportunity to intervene prior to malignant progression.<br />
Effective implementation of that strategy has been hampered by difficulties in clearly distinguishing cystic lesions based on<br />
differences in their malignant potential. In collaboration with Dr. Diane Simeone at the University of Michigan, we have identified<br />
glycan variants of secreted mucins that distinguish benign from pre-cancerous cysts with an 87% accuracy—better than the best<br />
current markers. Ongoing work is aimed at validating and building upon these results. Ultimately, we hope to implement a test<br />
that could be used to determine which pancreatic cysts should be surgically removed in order to prevent progression to cancer.<br />
Origin and function of secreted glycan alterations in pancreatic cancer<br />
Our laboratory also studies the origins and functions of cancer-cell secretions bearing altered glycans. The carbohydrate<br />
alterations observed in pancreatic tumors are strongly associated with accelerated disease progression, but it is not known<br />
whether these alterations functionally contribute to that progression. We have shown that certain glycoprotein alterations<br />
Figure 2<br />
are likely the product of subpopulations of tumor<br />
cells that are more likely to be aggressive. Using<br />
ALSA in a study of cultured pancreatic cancer<br />
cells, we have shown that cells bearing markers<br />
of high tumor-forming capability (termed “cancer<br />
stem cell markers”) display distinct glycan characteristics.<br />
The glycans of such cancer cells<br />
are distinctly altered in response to inflammatory<br />
signaling from the environment, showing the link<br />
between secreted glycan structures and the<br />
cellular state. Additional studies have shown<br />
distinct glycan alterations produced when cells<br />
transition from a stationary to a migratory state.<br />
This transition initiates metastasis and results in<br />
tumors at new sites. This work clearly links the<br />
origin of particular cancer-associated glycans with<br />
aggressive cancer cells.<br />
Figure 2. Distinct changes to glycan levels associated with cell type. Cell lines were treated with various pro-inflammatory signals,<br />
including oxidative stress (H 2<br />
O 2<br />
) and the cytokines IFNg, TNFa, or IL-a1. The cell lines and their treatments are indicated by the column<br />
labels. Six cell lines were treated: two bearing cell-surface markers characteristic of tumorigenicity (labeled in red); two not bearing the<br />
markers (labeled in black); and two partially bearing the markers (labeled in green). Using the ALSA assay, the levels of various glycans<br />
on the mucins MUC1, MUC5AC, and MUC16 in the secretions of the cells were measured before and after treatment. The row labels<br />
indicate the lectin used for detection (which determines the glycan detected) and the capture antibody. The color of each square<br />
represents the fold-change of the signal after treatment divided by the signal before treatment. The cells bearing markers of tumorigenicity<br />
uniquely increased particular glycans, showing a difference from the other cells in their glycan characteristics.<br />
28
VARI | <strong>2009</strong><br />
We are pursuing the hypothesis that the distinct glycans and glycoproteins secreted by aggressive or tumor-initiation cancer<br />
cells contribute to cancer progression through interactions with cells and proteins of the tumor environment. Evidence from our<br />
laboratory suggests that these secretions produce a higher state of inflammation and weaker immune recognition of the cancer<br />
cells. Our goals are to characterize the glycan alterations and their protein carriers that are unique to aggressive subsets of<br />
cancer cells and to understand the mechanisms by which these molecules affect host cells and promote tumor progression.<br />
In addition, we are investigating new strategies for treating cancer based on these observations. Targeting the functions of the<br />
aggressive subpopulations of cancer cells could be highly effective.<br />
External Collaborators<br />
Michelle Anderson, Philip Andrews, Dean Brenner, Irwin Goldstein, Venkat Keshamouni, Gilbert Omenn, and Diane Simeone,<br />
University of Michigan, Ann Arbor<br />
Randall Brand and Anna Lokshin, University of Pittsburgh, Pennsylvania<br />
William Catalona, Northwestern University, Evanston, Illinois<br />
Terry Du Clos, University of New Mexico, Albuquerque<br />
Ziding Feng and Samir Hanash, Fred Hutchinson Cancer Research Center, Seattle, Washington<br />
Weimin Gao, Texas Tech University, Lubbock<br />
William Hancock, Northeastern University, Boston, Massachusetts<br />
Michael A. Hollingsworth, University of Nebraska, Omaha<br />
Raju Kucherlapati, Harvard Medical School, Boston, Massachusetts<br />
Recent Publications<br />
Wu, Yi-Mi, and Brian. Haab. In press. The nature and function of glycan alterations in pancreatic cancer. In Drug Discovery in<br />
Pancreatic Cancer: Models and Techniques, Haiyong Han and Paul Grippo, eds. Springer Verlag.<br />
Hung, K.E., V. Faca, K. Song, D. Sarracino, L.G. Richard, B. Krastins, S. Forrester, A. Porter, A. Kunin, U. Mahmood,<br />
B.B. Haab, et al. <strong>2009</strong>. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with<br />
intestinal tumorigenesis. Cancer Prevention and Research 2(3): 224–233.<br />
Wu, Yi-Mi, D. David Novack, Gilbert S. Omenn and Brian B. Haab. <strong>2009</strong>. Mucin glycosylation is altered by pro-inflammatory<br />
signaling in pancreatic cancer cells. Journal of Proteome Research 8(4): 1876–1886.<br />
Yue, Tingting, and Brian B. Haab. <strong>2009</strong>. Microarrays in glycoproteomics research. Clinics in Laboratory Medicine<br />
29(1): 15–29.<br />
Chen, S., and B.B. Haab. 2008. Antibody microarrays for protein and glycan detection. In Clinical Proteomics, J. Van Eyk<br />
and M. Dunn, eds. Weinheim, Germany: Wiley-VCH.<br />
From left: Sinha, Antecki, Wu, Haab, Nelson, Maupin, VanOcker, Babins, Kluck, Yue<br />
29
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Jeffrey P. MacKeigan, Ph.D.<br />
Laboratory of Systems Biology<br />
Dr. MacKeigan received his Ph.D. in microbiology and immunology at the University of North Carolina<br />
Lineberger Comprehensive Cancer Center in 2002. He then served as a postdoctoral fellow in the<br />
laboratory of John Blenis in the Department of Cell Biology at Harvard Medical School. In 2004, he<br />
joined Novartis Institutes for Biomedical Research in Cambridge, Massachusetts, as an investigator<br />
and project leader in the Molecular and Developmental Pathways expertise platform. Dr. MacKeigan<br />
joined VARI in June 2006 as a <strong>Scientific</strong> Investigator.<br />
Staff Students Visiting Scientists<br />
Brendan Looyenga, Ph.D.<br />
Amy Nelson<br />
30<br />
Megan Goodall, B.S.<br />
Jon Karnes, B.S.<br />
Michael Shaheen, B.S.<br />
Katie Sian, B.S.<br />
Laura Westrate, B.S.<br />
Natalie Wolters, B.S.<br />
Cheri Ackerman<br />
James Hogan<br />
Bodour Salhia, Ph.D.<br />
Brad Brooks, Ph.D.
VARI | <strong>2009</strong><br />
Research Interests<br />
The primary focus of the Systems Biology laboratory is identifying and understanding the genes and signaling pathways that,<br />
when mutated, contribute to the pathophysiology of cancer. We take advantage of RNA interference (RNAi) and novel proteomic<br />
approaches to identify the enzymes that control cell growth, proliferation, and survival. For example, after screening the human<br />
genome for more than 600 kinases and 200 phosphatases—called the “kinome” and “phosphatome”, respectively—that act<br />
with chemotherapeutic agents in controlling apoptosis, we identified several essential kinases and phosphatases whose roles<br />
in cell survival were previously unrecognized. We are asking several questions. How are these survival enzymes regulated at<br />
the molecular level? What signaling pathway(s) do they regulate? Does changing the number of enzyme molecules present<br />
inhibit waves of compensatory changes at the cellular level (system-level changes)? What are the system-level changes after<br />
reduction or loss of each gene?<br />
Mitochondrial dysfunction in cancer<br />
Mitochondria are dynamic organelles that house many crucial cellular processes. While mitochondria are best known for<br />
producing more than 90% of cellular ATP and for releasing cytochrome c during apoptosis, they also modulate mitochondrial<br />
dynamics and ion homeostasis, oxidize carbohydrates and fatty acids, and participate in numerous other molecular signaling<br />
pathways. Disruption of mitochondrial function contributes to the etiology of at least fifty diseases, including cancer, underscoring<br />
the importance of identifying the molecular components that regulate normal and pathological function in these organelles.<br />
Similar to the discovery of the BCL-2<br />
Figure 1<br />
family members, which play key roles in<br />
mitochondrial apoptosis, the discovery<br />
of enzymes that regulate mitochondrial<br />
function (cytochrome c release, ATP production,<br />
and fission/fusion) will provide<br />
critical insights into the physiology of<br />
this organelle and how this physiology is<br />
disrupted in cancer.<br />
Figure 1. Mitochondrial dynamics as visualized by MitoTraker staining (red). As an outcome, mitochondrial dysfunction from<br />
a single kinase or phosphatase may have consequences that range from defects in energy metabolism to the etiology of complex<br />
diseases such as cancer. Our preliminary data with a mitochondrial kinase and two different mitochondrial phosphatases demonstrate<br />
that, when lost, the kinase decreases ATP production and drives mitochondrial fusion, while each phosphatase studied leads to an<br />
increase in ATP production. We have data that excessive or even modest increases in ATP levels may completely prevent mitochondrialdependent<br />
apoptosis. The significance of our work is that we have identified the specific mitochondrial signaling proteins that interact in<br />
a complex with key components of the electron transport chain and also with the mitochondrial fission/fusion machinery. Related to this,<br />
we have also identified a mitochondrial-specific kinase that controls the dynamic nature of the mitochondria, specifically mitochondrial<br />
fission and fusion.<br />
31
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Monitoring cellular signaling and autophagy<br />
Macroautophagy is a dynamic process whereby portions of the cytosol are encapsulated in double-membrane vesicles and<br />
delivered to a lysosome for degradation. Phosphatidylinositol-3-phosphate, or PI(3)P, is generated on the earliest autophagic<br />
membrane (phagophore) and recruits effector proteins needed for this process. The production of PI(3)P by the class III PI3-<br />
kinase Vps34 has been well established, but phosphatases that dephosphorylate this lipid during autophagy are unknown. To<br />
identify such enzymes, we screened human phosphatase genes by RNA interference (RNAi) and found that loss of a specific<br />
phosphatase in the human genome increases cellular PI(3)P and hyperactivates autophagy. This autophagic phenotype was<br />
confirmed in knock-out MEFs when compared with wild-type counterparts. Further, we discovered that this classically defined<br />
phosphatase harbors lipid phosphatase activity and its active site binds PI(3)P. Our findings suggest a novel role for these<br />
enzymes in cancer and provide insight into the regulation of autophagy. Mechanistic knowledge of this process is critical for<br />
understanding and targeting therapies for several human diseases, including Alzheimer disease and prostate cancer, in which<br />
abnormal autophagy may be pathological.<br />
Uncontrolled cellular survival and chemoresistance is a therapeutic problem that severely limits successful treatment of most<br />
human cancers. This is particularly true of colorectal cancer, in which the development of resistance is common: most anticancer<br />
regimens are ineffective, with the five-year survival rates for late-stage colorectal cancer being only 8%. How colorectal<br />
cancer resistance develops is largely unknown, and the response to therapy varies based on individual patient tumors. With<br />
this in mind, how can we prevent cancer emergence or progression at the level of individual tumors? Recent studies have<br />
shown that a large percentage of colorectal tumors have mutations in a key gene, for class I PI3 kinase. While mutations play<br />
an important causative role in colorectal cancer, it is currently unclear how these mutations can be exploited as drug targets<br />
and whether we can develop targeted cancer agents based on the gene. We have ongoing projects to determine the molecular<br />
pathways (and genes) that can be used to prevent the progression of precancerous lesions to colorectal cancer.<br />
Parkinson disease–associated genes in cancer<br />
Dysregulation of receptor tyrosine kinase signaling is a common oncogenic mechanism in human cancer. Abnormal activation<br />
of these receptors is associated with a loss of growth factor dependence, resulting in uncontrolled proliferation and<br />
survival of cancer cells. The receptor tyrosine kinase MET is often genetically amplified and overexpressed in human tumors.<br />
Because simple overexpression of MET is insufficient to mediate its activation, additional “hits” such as activating mutations<br />
or overexpression of its ligand, hepatocyte growth factor (HGF), often accompany MET genetic amplification. In the absence<br />
of these secondary events, it is not always clear how MET becomes activated and drives oncogenesis. We have identified a<br />
novel mechanism for MET activation that is driven by genetic co-amplification of a second protein kinase. The identification of<br />
alternative mechanisms that mediate MET activation in these instances is crucial for<br />
Figure 2<br />
the design of rationally targeted therapies aimed at interruption of oncogenic signaling<br />
in cancer. This project is a collaboration with VARI’s Kyle Furge, Bin Teh, and George<br />
Vande Woude.<br />
Figure 2. Depletion of specific kinases using RNAi decreases receptor tyrosine<br />
kinase activation. Global analysis of RTK phosphorylation in kidney cancer cells<br />
demonstrates significant activation of only EGFR (black arrow) and MET (white arrow) under<br />
basal conditions.<br />
32
VARI | <strong>2009</strong><br />
External Collaborators<br />
Dana Farber Cancer Institute, Boston, Massachusetts<br />
McGill Cancer Center, Montreal, Canada<br />
Michigan Medical, P.C., Grand Rapids<br />
Michigan State University, East Lansing<br />
Newcastle University, Newcastle upon Tyne, U.K.<br />
Novartis Institutes for Biomedical Research, Cambridge, Massachusetts<br />
Ontario Cancer Institute, Toronto, Canada<br />
Spectrum Health, Grand Rapids, Michigan<br />
St. Jude Children’s Hospital, Memphis, Tennessee<br />
Translational Genomics Research Institute, Phoenix, Arizona<br />
University of Virginia, Charlottesville<br />
Recent Publications<br />
Nicklin, Paul, Philip Bergman, Bailin Zhang, Ellen Triantafellow, Henry Wang, Beat Nyfeler, Haidi Yang, Marc Hild, Charles<br />
Kung, Christopher Wilson, et al. <strong>2009</strong>. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136(3):<br />
521–534.<br />
Looyenga, Brendan D., Alyse M. DeHaan, and Jeffrey P. MacKeigan. 2008. PINK1 (PARK6). UCSD-Nature Molecule Pages.<br />
June. doi:10.1038/mp.a003826.01. http://www.signaling-gateway.org/molecule/query?afcsid=A003826<br />
From left: Sian, Wolters, MacKeigan, Nelson, Looyenga, Goodall, Karnes, Westrate<br />
33
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Quantitative mass spectrometry: it’s all in the peaks!<br />
Liquid chromatography-mass spectrometry (LC-MS) is being widely used to identify, quantify, and compare hundreds, even thousands of<br />
proteins in diseased versus normal samples, with a goal of better understanding cancer systems biology and finding biomarkers that will improve<br />
clinical decision making. Shown in various colors are sections of mass spectra from LC-MS chromatograms of cachexia, metastatic, and<br />
non-metastatic disease models, demonstrating the fine detail of protein characterization that can be obtained from complex protein samples.<br />
Graphs courtesy of Greg Cavey.<br />
34
VARI | <strong>2009</strong><br />
Cindy K. Miranti, Ph.D.<br />
Laboratory of Integrin Signaling and Tumorigenesis<br />
Dr. Miranti received her M.S. in microbiology from Colorado State University in 1982 and her Ph.D. in<br />
biochemistry from Harvard Medical School in 1995. She was a postdoctoral fellow in the laboratory<br />
of Joan Brugge at ARIAD Pharmaceuticals, Cambridge, Massachusetts, from 1995 to 1997 and in<br />
the Department of Cell Biology at Harvard Medical School from 1997 to 2000. Dr. Miranti joined<br />
VARI as a <strong>Scientific</strong> Investigator in January 2000. She is also an Adjunct Assistant Professor in the<br />
Department of Physiology at Michigan State University and an Assistant Professor in the Van Andel<br />
Education Institute.<br />
Staff<br />
Electa Park, Ph.D.<br />
Kristin Saari, M.S.<br />
Lia Tesfay, M.S.<br />
Veronique Schulz, B.A.<br />
Students<br />
Jelani Zarif, M.S.<br />
Laura Lamb, B.S.<br />
Susan Spotts, B.S.<br />
Erica Bechtel<br />
Eric Graf<br />
Fraser Holleywood<br />
Gary Rajah, Jr.<br />
35
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
Our laboratory is interested in understanding the mechanisms by which integrin receptors, interacting with the extracellular<br />
matrix (ECM), regulate cell processes involved in the development and progression of cancer. Using tissue culture models,<br />
biochemistry, molecular genetics, and mouse models, we are defining the cellular and molecular events involved in integrindependent<br />
adhesion and downstream signaling that are important for prostate tumorigenesis and metastasis.<br />
Project 1: Integrin crosstalk in normal and tumor prostate epithelial cells<br />
In the human prostate gland, a3b1 and a6b4 integrins on epithelial cells bind to the ECM protein laminin 5 in the basement<br />
membrane. In tumor cells, however, the a3 and b4 integrin subunits disappear—as does laminin 5—and the tumor cells<br />
express primarily a6b1 and adhere to a basement membrane containing laminin 10. There is also an increase in the expression<br />
of the receptor tyrosine kinases EGFR and c-Met in tumor cells, and our laboratory has demonstrated that integrins cooperate<br />
with these receptors. Two fundamental questions in our lab are whether the changes in integrin and matrix interactions that<br />
occur in tumor cells are required for or help to drive the survival of tumor cells, and whether integrin cooperation with EGFR or<br />
c-Met is important for that cell survival.<br />
Integrins and RTKs in prostate epithelial cell survival<br />
By interacting with the ECM, integrins stimulate intracellular signaling transduction pathways that regulate cell shape, proliferation,<br />
migration, survival, gene expression, and differentiation. Integrins do not act autonomously, but “crosstalk” or cooperate<br />
with receptor tyrosine kinases (RTKs) to regulate many of these cellular processes. Published studies from our lab indicate<br />
that integrin-mediated adhesion to ECM proteins activates the epidermal growth factor receptors EGFR/ErbB2 and the HGF/<br />
SF receptor c-Met. We have shown that integrin-mediated activation of these receptors is ligand-independent and is required<br />
for integrin-mediated cell survival of prostate epithelial cells. However, the mechanisms by which the RTKs cooperate with<br />
integrins to regulate survival are different.<br />
The ability of EGFR to support integrin-mediated cell survival of normal primary prostate epithelial cells (PECs) on their<br />
endogenous matrix, laminin 5, is mediated through a3b1 integrin and requires signaling downstream to Erk. Disruption of<br />
this pathway leads to a caspase-independent mechanism of cell death resembling senescence/differentiation. On the other<br />
hand, loss of c-Met results in classic apoptotic cell death. Surprisingly, we found that c-Met regulates integrin-mediated<br />
survival by stabilizing a3b1 integrin expression and that regulation of integrin expression by c-Met occurs independently<br />
of its kinase activity. We are mapping the domains on c-Met that are required to rescue a3b1 integrin expression. The<br />
hypothesis being tested involves a potential scaffolding function of c-Met in suppressing the function of a cell surface death<br />
receptor called Fas and preventing the loss of a3b1 integrin and induction of death.<br />
Integrin control of the autophagy survival pathway<br />
During these studies, we also discovered that growth factor–deprived PECs adherent to laminin 5 robustly activate the autophagy<br />
survival pathway. Disruption of this pathway leads to apoptotic cell death, and a3b1 integrin is required for efficient autophagy<br />
induction. Because loss of c-Met reduces a3b1 integrin expression, autophagy induction is blocked in c-Met-inhibited cells.<br />
Preliminary data suggest that a3b1 integrin regulates the assembly of autophagosomes. Future work will be focused on<br />
identifying which molecules in the autophagy pathway are controlled by integrins. Our hypothesis is that under starvation<br />
conditions, integrins regulate the assembly of a FAK/FIP200 complex that controls autophagy.<br />
It is quite controversial as to whether inhibition or augmentation of autophagy is required for tumorigenesis and metastasis.<br />
Interestingly, immortalization of PECs or fibroblasts completely blocks the ability of autophagy-inducing stimuli to induce<br />
autophagy. In PECs immortalized by HPV E6/E7, we discovered a dramatic increase in PI-3K activity, which is known to inhibit<br />
autophagy via activation of mTor. However, blocking this pathway failed to restore the autophagic response. E7 is known to<br />
inhibit PP2A, and PP2A is required for autophagy induction in yeast downstream of mTor, but its role in mammalian cells has<br />
not been investigated. We will be following this line of investigation in both the virally immortalized PECs and in spontaneously<br />
immortalized cells. Ultimately, the effect of introducing prostate-specific oncogenes into PECs on the autophagy response will<br />
be analyzed.<br />
36
VARI | <strong>2009</strong><br />
Project 2: Integrin and AR relationships in prostate cancer<br />
All primary and metastatic prostate cancers express the androgen receptor (AR), and in late-stage disease it is often amplified<br />
or mutated. In the normal gland, the AR-expressing epithelial cells do not interact with the ECM in the basement membrane;<br />
however, all AR-expressing tumor cells do have such interactions. In normal cells, AR expression suppresses growth and<br />
promotes differentiation, but in tumor cells AR expression promotes cell growth and is required for cell survival. The mechanisms<br />
that lead to the change from growth inhibition and differentiation to growth promotion and survival are unknown. Our<br />
hypothesis is that adhesion to the ECM by the tumor cells is responsible for driving the change in AR function by initiating<br />
crosstalk between AR and integrins.<br />
AR and integrin-mediated survival signaling in prostate tumor cells<br />
Adhesion of PC3 metastatic prostate cancer cells to laminin and treatment with PI-3K inhibitors induces cell death. However,<br />
we found that reexpression of AR prevented that cell death in an androgen-independent manner. We have determined that<br />
AR expression results in increased expression of a6b1 integrin, the receptor for laminin. In addition, there is an increase in<br />
Bcl-xL levels. The increase in Bcl-xL is dependent on a6 integrin, and both integrin and Bcl-xL are dependent on AR. Thus,<br />
AR-expressing tumor cells are likely to survive better when they remain adherent to the laminin-rich ECM that is present in the<br />
prostate gland. Survival under these conditions appears to depend on the ability of AR to enhance expression of the laminin<br />
receptor, a6b1 integrin, which in turn stimulates Bcl-xL expression. These findings have broad implications for therapies<br />
specifically targeting the PI-3K pathway, in that AR-expressing cells may harbor an alternative survival pathway via integrins.<br />
We are currently determining how AR regulates the expression of a6 integrin and whether the transcriptional function of AR is<br />
required for the survival phenotype.<br />
AR-expressing cells also have elevated Src activity. Loss of Src did not impact cell survival, but these cells display increased<br />
cell adhesion, spreading, and migration. Future studies will be aimed at determining if these cells are also more aggressive in<br />
our in vivo metastasis models and if AR is responsible for controlling this. The survival signaling pathways observed in vitro will<br />
also be tested in our metastasis animal models.<br />
AR and integrin crosstalk in primary prostate epithelial cells<br />
Our ability to understand AR function in tumor cells relative to normal cells is hampered by the lack of a cell culture model in<br />
which normal cells naturally express AR. We sought to solve this problem by identifying the conditions necessary to induce<br />
the differentiation of normal human prostate basal epithelial cells (which do not express AR) into secretory AR-expressing<br />
cells. Combined treatment of confluent monolayers of human basal prostate epithelial cells with KGF and DHT stimulates the<br />
production of a second layer of cells, analogous to a stratified epithelium. The upper-layer cells express epithelial differentiation<br />
markers, AR, and AR-regulated genes, but no longer express integrins or basal cell markers. The upper secretory cell layer can<br />
easily be dissociated from the bottom basal cell layer and analyzed biochemically. Because integrins are no longer expressed<br />
in the secretory cells (as seen in vivo), we sought to determine how these cells survive. We found a dramatic increase in<br />
E-cadherin expression in the differentiated cells. The secretory AR-positive cells no longer rely on integrin or integrin-activated<br />
signaling pathways such as EGFR/c-Met/Erk; they now depend on E-cadherin and PI-3K signaling for their survival. Also, as<br />
has been demonstrated in in vivo models, these cells do not need androgen or AR for survival.<br />
Now that we have established a working differentiation model, we are poised to manipulate the cells by systematic introduction<br />
of oncogenic mutations known to be associated with the development of prostate cancer. As proof of concept, we are also<br />
capable of inducing the differentiation response in our immortalized PECs. Our hypothesis is that activation of oncogenes<br />
during differentiation will cause a dependence on AR for survival, which will elevate a6b1 integrin.<br />
Thus we have established two different models for studying AR in prostate cells. In both models the expression of AR has a<br />
major impact on integrin expression and function, indicating there is significant “crosstalk” between integrins and AR.<br />
37
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Project 3: Role of CD82 in prostate cancer metastasis<br />
Death from prostate cancer is due to the development of metastatic disease, which is difficult to control. CD82/KAI1 is a<br />
metastasis suppressor gene whose expression is specifically lost in metastatic cancer, but not in primary tumors. Interestingly,<br />
CD82/KAI1 (a member of the tetraspanin family) is known to associate with both integrins and receptor tyrosine kinases. Our<br />
goal has been to determine how loss of CD82/KAI1 expression promotes metastasis by studying the role of CD82/KAI1 in<br />
integrin and receptor tyrosine kinase crosstalk.<br />
Mechanism of CD82 suppression of c-Met<br />
We have found that reexpression of CD82/KAI1 in metastatic tumor cells suppresses laminin-specific migration and invasion<br />
via suppression of both integrin- and ligand-induced activation of c-Met. Thus, CD82/KAI1 normally acts to regulate signaling<br />
through c-Met; upon CD82 loss in tumor cells, signaling through c-Met is increased, leading to increased invasion. We<br />
are currently determining the mechanism by which CD82/KAI1 down-regulates c-Met signaling. So far our studies indicate<br />
that c-Met and CD82 do not directly interact, and CD82 may act to suppress c-Met signaling indirectly by dispersing the<br />
c-Met aggregates on metastatic tumor cells into monomers, thus blocking signaling. We have generated mutants of CD82 to<br />
determine which part of the CD82 molecule is required for suppression of c-Met activity. In addition, we have determined that<br />
reexpression of CD82 in tumor cells induces a physical association between CD82 and a related family member, CD9. Loss<br />
of CD9 prevents CD82 from suppressing c-Met. We are currently determining whether CD82/CD9 association with integrins<br />
is required to suppress c-Met.<br />
CD82 control of metastasis and c-Met activation in vivo<br />
We have initiated several mouse studies to determine the mechanism by which loss of CD82 promotes metastasis in vivo. The<br />
ability of DU145 prostate cancer cells to metastasize depends on activation of c-Met. Using transgenic SCID mice that express<br />
the human version of HGF (only human HGF will activate human c-Met), we have been able to demonstrate that DU145 tumor<br />
cells will metastasize only in the HGF/SCID mice, but not in regular SCID mice. Under these conditions, reexpression of CD82<br />
completely suppresses metastasis and there is a dramatic reduction in c-Met activity in the tumors. Mutants that no longer<br />
suppress c-Met activity in vitro will be used to demonstrate that they are also no longer capable of suppressing metastasis in<br />
the HGF/SCID mice.<br />
In addition, we have generated mice with conditional loss of CD82 expression in the prostate, as well as mice with complete<br />
CD82 loss in all tissues. These mice have been crossed with mice that develop only primary tumors (Pten conditional) in order<br />
to determine if the loss of CD82 is sufficient to induce prostate cancer metastasis. The mice are currently aging and being<br />
monitored for the development of tumors and metastases. Future studies will include back-crossing to alternative backgrounds<br />
and crossings with other tumor-prone mice.<br />
Recent Publications<br />
Miranti, C.K. <strong>2009</strong>. Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cellular Signalling<br />
21(2): 196–211.<br />
From left: Miranti, Schulz,<br />
Spotts, Lamb, Saari,<br />
Tesfay, Park, Zarif<br />
38
VARI | <strong>2009</strong><br />
James H. Resau, Ph.D.<br />
Division of Quantitative Sciences<br />
Laboratory of Analytical, Cellular, and Molecular Microscopy<br />
Laboratory of Microarray Technology<br />
Laboratory of Molecular Epidemiology<br />
Dr. Resau received his Ph.D. from the University of Maryland School of Medicine in 1985. He has been<br />
involved in clinical and basic science imaging and pathology-related research since 1972. Between<br />
1968 and 1994, he was in the U.S. Army (active duty and reserve assignments) and served in Vietnam.<br />
From 1985 until 1992, Dr. Resau was a tenured faculty member at the University of Maryland School of<br />
Medicine, Department of Pathology. Dr. Resau was the Director of the Analytical, Cellular and Molecular<br />
Microscopy Laboratory in the Advanced BioScience Laboratories–Basic Research Program at the<br />
National Cancer Institute, Frederick Cancer Research and Development Center, Maryland, from 1992<br />
to 1999. He joined VARI as a Special Program Senior <strong>Scientific</strong> Investigator in June 1999 and in 2003<br />
was promoted to Deputy Director. In 2004, Dr. Resau assumed as well the direction of the Laboratory<br />
of Microarray Technology to consolidate the imaging and quantification of clinical samples in a CLIAtype<br />
research laboratory program. In 2005, Dr. Resau was made the Division Director of the quantitative<br />
laboratories (pathology-histology, microarray, proteomics, epidemiology, and bioinformatics), and in<br />
2006 he was promoted to Distinguished <strong>Scientific</strong> Investigator.<br />
Staff<br />
Students<br />
Visiting Scientist<br />
Eric Kort, M.D.<br />
Sok Kean Khoo, Ph.D.<br />
Brendan Looyenga, Ph.D.<br />
Bree Berghuis, B.S., HTL<br />
(ASCP), QIHC<br />
Eric Hudson, B.S.<br />
Angie Jason, B.S.<br />
Natalie Kent, B.S.<br />
Ken Olinger, B.S.<br />
Kristin VandenBeldt, B.S.<br />
JC Goolsby, A.A.<br />
Danielle Burgenske, B.S.<br />
Kevin Coalter, B.S.<br />
Pete Haak, B.S.<br />
Kimberly Paquette, B.S.<br />
Sarah Barney<br />
Janell Carruthers<br />
Katsuo Hisano<br />
Rebecca O’Leary<br />
Sara Ramirez<br />
Allison Vander Ploeg<br />
Katie Van Drunen<br />
Yair Andegeko<br />
39
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
The Division of Quantitative Sciences includes the laboratories of Analytical, Cellular, and Molecular Microscopy (ACMM),<br />
Microarray Technology, Computational Biology, Molecular Epidemiology, and Mass Spectrometry and Proteomics. The Division’s<br />
laboratories use objective measures to define pathophysiologic events and processes.<br />
The ACMM laboratory prepares samples by either paraffin or frozen methods and has programs in pathology, histology, and<br />
imaging to describe and visualize changes in cell, tissue, or organ structure. Our imaging instruments allow us to visualize<br />
cells and their components with striking clarity, and they enable researchers to determine where in a cell particular molecules<br />
are located. An archive of pathology tissues in paraffin blocks (Van Andel Tissue Repository, or VATR) is being accumulated<br />
with the cooperation of local hospitals. The archive currently has approximately 250,000 paraffin blocks representing 150,000<br />
cases. In collaboration with Tom Barney from VAI-IT, clinical data is being added into VATR for hundreds of the samples each<br />
week by digital parsing of pathology report texts sent electronically from the hospital files. VATR is used to track samples<br />
coming from the hospitals, along with all of the data and images generated from research. Images from the Aperio ScanScope<br />
are automatically imported into VATR and associated with the appropriate sample. The ACMM lab also carries out research<br />
that will improve our ability to quantify images. We are able to image using either fluorescent (e.g., FITC, GFP) or chromatic<br />
agents (e.g., DAB, H&E) and separate the components using our confocal, Nuance, or Maestro instruments.<br />
The Laboratory of Microarray Technology provides gene expression arrays, miRNA arrays, and array CGH using the Agilent<br />
microarray platform and cDNA platform capability. Samples can be prepared from a variety of species. Genomic DNA or total<br />
RNA from a wide range of tissues including blood and fresh or frozen tissues have been analyzed. A recent gene expression<br />
discovery was made using archived newborn blood spots, in collaboration with Dr. Nigel Paneth at MSU. We showed that<br />
thousands of gene signatures can be obtained using low-resolution gene expression arrays, enabling clinical research into<br />
the origins, epidemiology, and diagnosis of human pediatric diseases. Feature extraction software reads and processes the<br />
raw microarray image files in an automated mode. Application-specific QC reports summarize the results and provide an<br />
accurate quality assessment. The output files are compatible with statistical analysis packages such as R and GeneSpring.<br />
Microarray technology plays an important part in the discovery of genetic signatures, copy number variations, and biomarkers<br />
for therapeutic purposes.<br />
Hauenstein Parkinson’s Center<br />
Throughout 2008 we continued our collaboration with the Hauenstein Parkinson’s Center, collecting blood samples and control<br />
samples from 216 individuals. Mutations/polymorphisms in the NR4A2 gene are being studied by DNA sequence analysis,<br />
motivated by our previous identification of this gene in a genomic screen for neuroprotective factors. We are particularly<br />
interested in polymorphisms in the DNA-binding domain of NR4A2, as changes to this region of the gene are most likely to<br />
affect its function.<br />
Identification of novel Parkinson-modifying genes with siRNA screening<br />
Small interfering RNA (siRNA) technology allows the specific knockdown of any mRNA/protein pair. Under the direction of<br />
VARI’s Jeff MacKeigan, postdoctoral fellow Brendan Looyenga has begun to use a subset of the siRNA library developed by<br />
Qiagen to individually target several classes of enzymes having pharmaceutical potential. Specifically, we are continuing a<br />
project to identify molecules that attenuate oxidative stress–induced toxicity in dopaminergic neurons; our initial focus is on<br />
phosphatases and kinases. We are validating the initial screening studies and we hope to extend these studies to include<br />
nuclear hormone receptors and G protein–coupled receptors.<br />
40
VARI | <strong>2009</strong><br />
Mouse models of Parkinson disease<br />
James Resau and Brendan Looyenga, in collaboration with VARI’s Bart Williams, are generating novel rodent models of dopaminergic<br />
cell loss in the brain using the DAT-cre mouse line, which specifically expresses the cre recombinase in dopaminergic<br />
neurons of the brain. Projects based on the DAT-cre mouse model include the following.<br />
• Imaging and isolation of primary dopaminergic neurons from mouse brain. Brendan Looyenga is continuing a<br />
project to generate mice that specifically express the LacZ reporter gene in dopaminergic neurons. With these<br />
mice we will assess the effect of cytotoxic agents (e.g., MMTP, rotenone, or 6-hydroxydopamine) on the number<br />
of dopaminergic cells, and more importantly, assess the ability of mice to recover from these insults. The DAT-cre/<br />
ROSA26 mice will also provide a source of highly pure dopaminergic neurons for in vitro studies.<br />
• Functional roles of the phosphatase PTEN in dopaminergic neurons. The phosphatase PTEN is a crucial signaling<br />
node in mammalian cells. PTEN catalyzes the removal of 3´ phosphates from phosphoinositol (PI), effectively<br />
antagonizing the activity of PI3-kinase. Loss of PTEN results in constitutively elevated levels of the phospholipids<br />
PI(3,4)P 2<br />
and PI(3,4,5)P 3<br />
, which strongly activate downstream effectors including AKT/PKB and mTOR. Hyperactive<br />
AKT is traditionally associated with cell survival and proliferation, while hyperactivation of mTOR is associated<br />
with cellular hypertrophy. Interestingly, these two effects often occur in mutually exclusive fashion depending on<br />
the status of the cell in which PTEN is deleted. Terminally differentiated cells usually display hypertrophy, and they<br />
rarely reenter the cell cycle upon PTEN deletion.<br />
To confirm that the DAT-cre mice only express the cre recombinase in terminally differentiated cells, we have crossed them<br />
to mice bearing a conditional knock-out allele for PTEN (PTEN loxP ). As expected, dopaminergic neurons in DAT-cre/PTEN loxP<br />
mice demonstrate constitutive activation of AKT and mTOR; however, they fail to develop neuronal tumors, suggesting that the<br />
loss of PTEN in dopaminergic neurons does not induce hyperproliferation. These cells do appear larger, consistent with the<br />
induction of hypertrophy. We are currently quantifying these observations for publication.<br />
We are also using the DAT-cre/PTEN loxP mice to elucidate the connection between PTEN and mitochondrial function. This<br />
connection is based on the ability of PTEN to increase expression of the familial Parkinson gene, PINK1, in cultured tumor cells.<br />
Several studies have shown that PINK1 plays a crucial role in the maintenance of mitochondrial fission/fusion homeostasis,<br />
implying that PTEN indirectly regulates mitochondrial function by controlling cellular PINK1 levels. To determine whether PTEN<br />
regulates PINK1 and mitochondrial function in normal cells, we are analyzing PINK1 expression levels and mitochondrial<br />
function in the dopaminergic neurons of DAT-cre/PTEN loxP mice. We hypothesize that loss of PTEN in these cells will result<br />
in decreased PINK1 expression, imbalances in mitochondrial fission/fusion, and increased oxidative stress. Studies in brain<br />
tissues and cultured primary neurons are ongoing.<br />
Other highlights<br />
Our GRAPCEP mentorship program continues for the ninth year and is now funded by Schering Plough. In 2008 we had two<br />
students from GRAPCEP, several undergraduate summer interns, and a graduate school student rotation.<br />
41
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Recent Publications<br />
Knudsen, Beatrice S., Ping Zhao, James Resau, Sandra Cottingham, Ermanno Gherardi, Eric Xu, Bree Berghuis, Jennifer<br />
Daugherty, Tessa Grabinski, Jose Toro, et al. <strong>2009</strong>. A novel multipurpose monoclonal antibody for evaluating human c-Met<br />
expression in preclinical and clinical settings. Applied Immunohistochemistry and Molecular Morphology 17(1): 56–67.<br />
Kort, Eric, Paul Norton, Peter Haak, Bree Berghuis, Sara Ramirez, and James Resau. <strong>2009</strong>. Gene expression profiling<br />
in veterinary and human medicine: overview of applications and proposed quality control practices. Veterinary Pathology,<br />
46(4): 598–603.<br />
Golan, Maya, Amnon Hizi, James H. Resau, Neora Yaal-Hahoshen, Hadar Reichman, Iafa Keydar, and Ilan Tsarfaty.<br />
2008. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia<br />
10(6): 521–533.<br />
Haak, Peterson T., Julia V. Busik, Eric J. Kort, Maria Tikhonenko, Nigel Paneth, and James H. Resau. 2008. Archived unfrozen<br />
neonatal blood spots are amenable to quantitative gene expression analysis. Neonatology 95(3): 210–216.<br />
Lindemann, Kristina, Nadia Harbeck, Ernst Lengyel, and James H. Resau. 2008. A special key for unlocking the door to<br />
targeted therapies of breast cancer. The <strong>Scientific</strong> World Journal 8: 905–908.<br />
Xie, Qian, Ryan Thompson, Kim Hardy, Lisa DeCamp, Bree Berghuis, Robert Sigler, Beatrice Knudsen, Sandra Cottingham,<br />
Ping Zhao, Karl Dykema, et al. 2008. A highly invasive human glioblastoma pre-clinical model for testing therapeutics.<br />
Journal of Translational Medicine 6: 77.<br />
From left: Resau, Kort, Haak, Goolsby, Khoo, Jason, Kent, VandenBeldt, Paquette, Hudson, Carruthers, Ramirez, Berghuis<br />
42
VARI | <strong>2009</strong><br />
Pamela J. Swiatek, Ph.D., M.B.A.<br />
Laboratory of Germline Modification and Cytogenetics<br />
Dr. Swiatek received her M.S. (1984) and Ph.D. (1988) degrees in pathology from Indiana University.<br />
From 1988 to 1990, she was a postdoctoral fellow at the Tampa Bay Research Institute. From<br />
1990 to 1994, she was a postdoctoral fellow at the Roche Institute of Molecular Biology in the<br />
laboratory of Tom Gridley. From 1994 to 2000, Dr. Swiatek was a research scientist and Director of<br />
the Transgenic Core Facility at the Wadsworth Center in Albany, N.Y., and an Assistant Professor in<br />
the Department of Biomedical Sciences at the State University of New York at Albany. She joined<br />
VARI as a Special Program Investigator in August 2000. She was the chair of the Institutional Animal<br />
Care and Use Committee from 2002 to 2008, and she is an Adjunct Assistant Professor in the<br />
College of Veterinary Medicine at Michigan State University. Dr. Swiatek received her M.B.A. in 2005<br />
from Krannert School of Management at Purdue University, and in 2006 she was promoted to Senior<br />
<strong>Scientific</strong> Investigator.<br />
Staff<br />
Sok Kean Khoo, Ph.D.<br />
Kellie Sisson, B.S.<br />
Laura Mowry, B.S.<br />
Julie Koeman, B.S., CLSp(CG)<br />
Diana Lewis<br />
Student<br />
Juraj Zahatnansky, B.S.<br />
43
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
The Germline Modification and Cytogenetics lab is a full-service lab that functions at the levels of service, research, and<br />
teaching to develop, analyze, and maintain mouse models of human disease. Our lab applies a business philosophy to core<br />
service offerings for both the VARI community and external entities. Our mission is to support mouse model and cytogenetics<br />
research with scientific innovation, customer satisfaction, and service excellence.<br />
Gene targeting<br />
Mouse models are produced using gene-targeting technology, a well-established, powerful method for inserting specific<br />
genetic changes into the mouse genome. The resulting mice can be used to study the effects of these changes in the complex<br />
biological environment of a living organism. The genetic changes can include the introduction of a gene into a specific site in<br />
the genome (gene “knock-in”) or the inactivation of a gene already in the genome (gene “knock-out”). Since these mutations<br />
are introduced into the reproductive cells known as the germline, they can be used to study the developmental aspects of gene<br />
function associated with inherited genetic diseases.<br />
The germline modification lab can also produce mouse models in which the gene of interest is inactivated in a target organ<br />
or cell line instead of in the entire animal. These models, known as conditional knock-outs, are particularly useful in studying<br />
genes that, if missing, cause the mouse to die as an embryo. The lab can produce mutant embryos that have a wild-type<br />
placenta using tetraploid embryo technology, which is useful when the gene-targeted mutation prevents implantation of the<br />
mouse embryo in the uterus. We also assist in the development of embryonic stem (ES) or fibroblast cell lines from mutant<br />
embryos, to allow for in vitro studies of the gene mutation.<br />
Our gene-targeting service encompasses three major procedures: DNA electroporation, clone expansion and cryopreservation,<br />
and microinjection. Gene targeting is initiated by mutating the genomic DNA of interest and inserting it into ES cells via<br />
electroporation. The mutated gene integrates into the genome and, by a process called homologous recombination, replaces<br />
one of the two wild-type copies of the gene in the ES cells. Clones are identified, isolated, and cryopreserved, and genomic<br />
DNA is extracted from each clone and delivered to the client for analysis. Correctly targeted ES cell clones are thawed,<br />
established into tissue culture, and cryopreserved in liquid nitrogen. Gene-targeting mutations are introduced by microinjection<br />
of the pluripotent ES cell clones into 3.5-day-old mouse embryos (blastocysts). These embryos, containing a mixture of<br />
wild-type and mutant ES cells, develop into mice called chimeras. The offspring of chimeras that inherit the mutated gene are<br />
heterozygotes possessing one copy of the mutated gene. The heterozygous mice are bred together to produce “knock-out<br />
mice” that completely lack the normal gene and have two copies of the mutant gene.<br />
Embryo/sperm cryopreservation<br />
We provide cryopreservation services for archiving and reconstituting valuable mouse strains. These cost-effective procedures<br />
decrease the need to continuously breed valuable mouse models, and they provide added insurance against the loss of custom<br />
mouse lines due to disease outbreak or a catastrophic event. Mouse embryos at various stages of development, as well as<br />
mouse sperm, can be cryopreserved and stored in liquid nitrogen; they can be thawed and used, respectively, by implantation<br />
into the oviducts of recipient mice or by in vitro fertilization of oocytes.<br />
44
VARI | <strong>2009</strong><br />
Cytogenetics<br />
Our lab also directs the VARI cytogenetics core, which uses advanced molecular techniques to identify structural and numerical<br />
chromosomal aberrations in mouse, rat, and human cells. Tumor, fibroblast, blood, or ES cells can be grown in tissue culture,<br />
growth-arrested, fixed, and spread onto glass slides. Karyotyping of chromosomes using Leishman- or Giemsa-stained<br />
(G-banded) chromosomes is our basic service; spectral karyotyping (SKY) analysis of metaphase chromosome spreads in 24<br />
colors can aid in detecting subtle and complex chromosomal rearrangements. Fluorescence in situ hybridization (FISH) analysis,<br />
using indirectly or directly labeled bacterial artificial chromosome (BAC) or plasmid probes, can also be performed on metaphase<br />
spreads or on interphase nuclei derived from tissue touch preps or nondividing cells. Sequential staining of identical metaphase<br />
spreads using FISH and SKY can help identify the integration site of a randomly integrated transgene. Recently, FISH has been<br />
widely used to validate microarray data by confirming amplification/gain or deletion/loss of chromosomal regions of interest.<br />
Speed congenics<br />
Congenic mouse strain development traditionally involves a series of backcrosses, transferring a targeted mutation or genetic<br />
region of interest from a mixed genetic donor background to a defined genetic recipient background (usually an inbred strain).<br />
This process requires about ten generations (2.5 to 3 years) to attain 99.9% of the recipient’s genome. Since congenic mice<br />
have a more defined genetic background, phenotypic characteristics are less variable and the effects of modifier genes can be<br />
more pronounced.<br />
Speed congenics, also called marker-assisted breeding, uses DNA markers in a progressive breeding selection to accelerate<br />
the congenic process. For high-throughput genotyping, we use the state-of-the-art Sentrix BeadChip technology from Illumina,<br />
which contains 1,449 mouse single nucleotide polymorphisms (SNPs). These SNPs are strain-specific and cover the 10 most<br />
commonly used inbred mouse strains for optimal marker selection. The client provides the genomic DNA of male mice from<br />
the second, third, and fourth backcross generations for genotyping. The males having the highest percentage of the recipient’s<br />
genome from each generation are identified, and these mice are bred by the client. Using speed congenics, 99.9% of congenicity<br />
can be achieved in five generations (about 1.5 years).<br />
Recent Publications<br />
Graveel, C.R., J.D. DeGroot, Y. Su, J.M. Koeman, K. Dykema, S. Leung, J. Snider, S.R. Davies, P.J. Swiatek, S. Cottingham,<br />
et al. <strong>2009</strong>. Met induces diverse breast carcinomas in mice and is associated with human basal breast cancer. Proceedings of the<br />
National Academy of Sciences U.S.A. 106(31): 12909–12914.<br />
From left: Swiatek, Sisson, Mowry, Lewis, Koeman<br />
45
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Astrocytes in mouse retina<br />
An epifluorescent image of a mouse retina at postnatal day 7, showing astrocytes stained green with glial fibrillary acidic protein (GFAP, which<br />
measures activated glial cells) and endothelial blood vessel cells stained red with GSA lectin. The image shows the intricate guidance pattern<br />
between the astrocytes and the vasculature in the superficial layer of the retina. Red dots are sprouting blood vessels seen in cross section as<br />
they penetrate to other retinal layers. Original magnification, 400×. Photo by Jennifer Bromberg-White of the Duesbery lab.<br />
46
VARI | <strong>2009</strong><br />
Bin T. Teh, M.D., Ph.D.<br />
Laboratory of Cancer Genetics<br />
Dr. Teh obtained his M.D. from the University of Queensland, Australia, in 1992, and his Ph.D. from<br />
the Karolinska Institute, Sweden, in 1997. Before joining the Van Andel Research Institute, he was an<br />
Associate Professor of Medical Genetics at the Karolinska Institute. Dr. Teh joined VARI as a Senior<br />
<strong>Scientific</strong> Investigator in January 2000. His research mainly focuses on kidney cancer, and he is<br />
currently on the Medical Advisory Board of the Kidney Cancer Association. Dr. Teh was promoted to<br />
Distinguished <strong>Scientific</strong> Investigator in 2005.<br />
Staff<br />
Students<br />
Eric Kort, M.D.<br />
Jindong Chen, Ph.D.<br />
Yan Ding, Ph.D.<br />
Vanessa Fogg, Ph.D.<br />
Dan Huang, Ph.D.<br />
Aikseng Ooi, Ph.D.<br />
David Petillo, Ph.D.<br />
Zhongfa (Jacob) Zhang, Ph.D.<br />
Mario Lucia, B.S.<br />
Sabrina Noyes, B.S.<br />
Doug Roossien Jr., B.S.<br />
Bill Wondergem, B.S.<br />
Mike Avallone<br />
Kristin Buzzitta<br />
Jim Fitzgerald<br />
John Snider<br />
47
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
Kidney cancer, or renal cell carcinoma (RCC), is the tenth most common cancer in the United States (51,000 new cases and<br />
more than 13,000 deaths a year). Its incidence has been increasing, a phenomenon that cannot be accounted for by the wider<br />
use of imaging procedures. We have established a comprehensive and integrated kidney research program, and our major<br />
research goals are 1) to identify the molecular signatures of different subtypes of kidney tumors, both hereditary and sporadic,<br />
and to understand how genes function and interact in giving rise to the tumors and their progression; 2) to identify and develop<br />
diagnostic and prognostic biomarkers for kidney cancer; 3) to identify and study novel and established molecular drug targets<br />
and their sensitivity and resistance; and 4) to develop animal models for drug testing and preclinical bioimaging.<br />
Our program to date has established a worldwide network of collaborators; a tissue bank containing fresh-frozen tumor pairs<br />
(over 1,500 cases) and serum; and a gene expression profiling database of 700 tumors, with long-term clinical follow-up<br />
information for half of them. Our program includes molecular subclassification using microarray gene expression profiling and<br />
bioinformatic analysis, generation of RCC mouse models, and more recently, molecular therapeutic studies.<br />
RCC genomics<br />
We have been using high-density single nucleotide polymorphism (SNP) arrays to genotype RCC samples, and by combing<br />
this data and the gene expression data (see below), we have identified candidate chromosomal regions and genes that are<br />
involved in different subsets of tumors.<br />
Gene expression profiling and bioinformatics<br />
To date, we have studied over 600 RCC specimens. We are currently focusing on analysis and data mining. Clinically, we<br />
continue to subclassify the tumors by correlation with clinicopathological information, including rarer forms of RCC such as<br />
translation-related papillary RCC, mixed epithelial and stromal tumors, and adult Wilms. We are also in the process of trying to<br />
understand the underlying molecular signatures of some of the so-called unclassified group of tumors for which the histological<br />
diagnosis is “unknown”. Our database has proven to be very useful in RCC research, since we can obtain differential expression<br />
data for any gene in seconds.<br />
Mouse models of kidney cancer and molecular therapeutic studies<br />
We have generated several kidney-specific conditional knock-outs including BHD, PTEN, and VHL. The first two knock-outs<br />
give rise to renal cysts and tumors including urothelial cancer of the renal pelvis, whereas the VHL knock-out remains neoplasiafree;<br />
double knock-outs are also being studied. We have successfully generated nine xenograft RCC models via subcapsular<br />
injection that have characteristic clinical features and outcomes. Tumors and serum have been harvested for a baseline data<br />
set. We are currently performing in vitro and in vivo studies on several new drugs for kidney cancer.<br />
48
VARI | <strong>2009</strong><br />
Targeted therapeutic studies<br />
We have focused on several targets we identified using gene expression profiling studies. In vitro and in vivo studies are being<br />
performed to verify these targets and their role in therapeutics. These include cell-cycle, proliferation, and migration assays<br />
to assess the cellular effects of these genes. In vivo studies are performed to understand the involvement of blood vessels in<br />
drug response.<br />
External Collaborators<br />
We have extensive collaborations with researchers and clinicians in the United States and overseas.<br />
Recent Publications<br />
Furge, K., and B.T. Teh. In press. Genetics of sporadic renal cell carcinoma. In Renal Cell Carcinoma, B.I. Rini and<br />
S.C. Campbell, eds. Clinical Oncology series, BC Decker Inc.<br />
Van Haaften, G., G.L. Dalgliesh, H. Davies, G. Bigness, C. Greenman, S. Edkins, C. Hardy, S. O’Meara, L. Chen, J. Teague,<br />
et al. In press. Somatic mutations of the histone H3K27 demethylase, UTX, in human cancer. Nature Genetics.<br />
Howell, Viive M., Anthony Gill, Adele Clarkson, Anne E. Nelson, Robert Dunne, Leigh W. Delbridge, Bruce G. Robinson,<br />
Bin T. Teh, Oliver Gimm, and Deborah J. Marsh. <strong>2009</strong>. Accuracy of combined protein gene product 9.5 and parafibromin<br />
markers for immunohistochemical diagnosis of parathyroid carcinoma. Journal of Clinical Endocrinology & Metabolism<br />
94(2): 434–441.<br />
Hui, Zhouguang, Maria Tretiakova, Zhongfa Zhang, Yan Li, Xiaozhen Wang, Julie Xiaohong Zhu, Yuanhong Gao, Weiyuan Mai,<br />
Kyle Furge, Chao-Nan Qian, et al. <strong>2009</strong>. Radiosensitization by inhibiting STAT1 in renal cell carcinoma. International Journal<br />
of Radiation Oncology Biology Physics 73(1): 288–295.<br />
Macher-Goeppinger, Stephan, Sebastian Aulmann, Katrin E. Tagscherer, Nina Wagener, Axel Haferkamp, Roland Penzel,<br />
Antje Brauckhoff, Markus Hohenfellner, Jaromir Sykora, Henning Walczak, et al. <strong>2009</strong>. Prognostic value of tumor<br />
necrosis factor–related apoptosis-inducing ligand (TRAIL) and TRAIL receptors in renal cell cancer. Clinical Cancer Research<br />
15(2): 650–659.<br />
Wang, Y., O. Roche, M.S. Yan, G. Finak, A.J. Evans, J.L. Metcalf, B.E. Hast, S.C. Hanna, B. Wondergem, K.A. Furge, et al.<br />
<strong>2009</strong>. Regulation of endocytosis via the oxygen-sensing pathway. Nature Medicine 15(3): 319–324.<br />
Zhou, Ming, Eric Kort, Philip Hoekstra, Michael Westphal, Cristina Magi-Galluzzi, Linda Sercia, Brian Lane, Brian Rini,<br />
Ronald Bukowski, and Bin T. Teh. <strong>2009</strong>. Adult cystic nephroma and mixed epithelial and stromal tumor of the kidney are<br />
the same disease entity: molecular and histological evidence. American Journal of Surgical Pathology 33(1): 72–80.<br />
Camparo, Philippe, Viorel Vasiliu, Vincent Molinié, Jerome Couturier, Karl J. Dykema, David Petillo, Kyle A. Furge,<br />
Eva M. Comperat, Marick Laé, Raymonde Bouvier, et al. 2008. Renal translocation carcinomas: clinicopathologic, immunohistochemical,<br />
and gene expression profiling analysis of 31 cases with a review of the literature. American Journal of Surgical<br />
Pathology 32(5): 656–670.<br />
49
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Chen, Jindong, Kunihiko Futami, David Petillo, Jun Peng, PengFei Wang, Jared Knol, Yan Li, Sok Kean Khoo, Dan Huang,<br />
Chao-Nan Qian, et al. 2008. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal neoplasia.<br />
PLoS One 3(10): e3581.<br />
Farber, Leslie, and Bin Tean Teh. 2008. CDC73 (cell division cycle 73, Paf1/RNA polymerase II complex<br />
component, homolog (S. cerevisiae)). Atlas of Genetics and Cytogenetics in Oncology and Haematology. February.<br />
http://AtlasGeneticsOncology.org/Genes/CDC73D181ch1q31.html.<br />
Koeman, Julie M., Ryan C. Russell, Min-Han Tan, David Petillo, Michael Westphal, Katherine Koelzer, Julie L. Metcalf,<br />
Zhongfa Zhang, Daisuke Matsuda, Karl J. Dykema, et al. 2008. Somatic pairing of chromosome 19 in renal oncocytoma is<br />
associated with deregulated EGLN2-mediated oxygen-sensing response. PLoS Genetics 4(9): e1000176.<br />
Kort, Eric J., Leslie Farber, Maria Tretiakova, David Petillo, Kyle A. Furge, Ximing J. Yang, Albert Cornelius, and Bin T. Teh. 2008.<br />
The E2F3–Oncomir-1 axis is activated in Wilms’ tumor. Cancer Research 68(11): 4034–4038.<br />
Matsuda, Daisuke, Sok Kean Khoo, Aaron Massie, Masatsugu Iwamura, Jindong Chen, David Petillo, Bill Wondergem,<br />
Michael Avallone, Stephanie J. Kloostra, Min-Han Tan, et al. 2008. Identification of copy number alterations and its association<br />
with pathological features in clear cell and papillary RCC. Cancer Letters 272(2): 260–267.<br />
Sarquis, Marta S., Leticia G. Silveira, Flavio J. Pimenta, Eduardo P. Dias, Bin T. Teh, Eitan Friedman, Ricardo S. Gomez,<br />
Gabriela C. Tavares, Charis Eng, and Luiz De Marco. 2008. Familial hyperparathyroidism: surgical outcome after 30 years<br />
follow-up in three families with germline HRPT2 mutations. Surgery 143(5): 630–640.<br />
Zhang, Zhong-Fa, Daisuke Matsuda, Sok Kean Khoo, Kristen Buzzitta, Elizabeth Block, David Petillo, Stéphane Richard,<br />
John Anema, Kyle A. Furge, and Bin T. Teh. 2008. A comparison study reveals important features of agreement and disagreement<br />
between summarized DNA and RNA data obtained from renal cell carcinoma. Mutation Research 657(1): 77–83.<br />
From left: Fogg, Ooi, Noyes, Ding, Zhang, Chen, Huang, Lucia, Petillo, Wondergem, Teh, Roossien<br />
50
VARI | <strong>2009</strong><br />
Steven J. Triezenberg, Ph.D.<br />
Laboratory of Transcriptional Regulation<br />
Dr. Triezenberg received his bachelor’s degree in biology and education at Calvin College in Grand<br />
Rapids, Michigan. His Ph.D. training in cell and molecular biology at the University of Michigan was<br />
followed by postdoctoral research in the laboratory of Steven L. McKnight at the Carnegie Institution<br />
of Washington. Dr. Triezenberg was a faculty member of the Department of Biochemistry and<br />
Molecular Biology at Michigan State University for more than 18 years, where he also served as<br />
associate director of the Graduate Program in Cell and Molecular Biology. In 2006, Dr. Triezenberg<br />
was recruited to VAI to serve as the founding Dean of the Van Andel Institute Graduate School and<br />
as a <strong>Scientific</strong> Investigator in the Van Andel Research Institute. He succeeded Gordon Van Harn as<br />
the Director of the Van Andel Education Institute in January <strong>2009</strong>.<br />
Staff<br />
Xu Lu, Ph.D.<br />
Jennifer Klomp, M.S.<br />
Glen Alberts, B.S.<br />
Carol Rappley<br />
Students<br />
Sebla Kutluay, B.S.<br />
Tim Caldwell<br />
Justyne Matheny<br />
Marian Testori<br />
51
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
The genetic information encoded in DNA must first be copied, in the form of RNA, before it can be translated into the proteins<br />
that do most of the work in a cell. Some genes must be expressed more or less constantly throughout the life of any eukaryotic<br />
cell. Others must be turned on (or turned off) in particular cells either at specific times or in response to a specific signal or<br />
event. Thus, regulation of gene expression is a key determinant of cell function. Our laboratory explores the mechanisms that<br />
regulate the first step in that flow, the process termed transcription.<br />
Over the past 20 years, my laboratory has used infection by herpes simplex virus as an experimental context for exploring the<br />
mechanisms of transcriptional activation. In the past 10 years, we have also asked similar questions in a very different biological<br />
context, the acclimation of plants to cold temperature.<br />
Transcriptional activation during herpes simplex virus infection<br />
Herpes simplex virus type 1 (HSV-1) causes the common cold sore or fever blister. The initial lytic (or productive) infection<br />
by HSV-1 results in the obvious symptoms in the skin and mucosa, typically in or around the mouth. After the initial infection<br />
resolves, HSV-1 finds its way into nerve cells, where the virus can hide in a latent mode for long times—essentially for the<br />
lifetime of the host organism. Occasionally, some trigger event (such as emotional stress, damage to the nerve from a sunburn,<br />
or a root canal operation) will cause the latent virus to reactivate, producing new viruses in the nerve cell and sending those<br />
viruses back to the skin to cause a recurrence of the cold sore.<br />
The DNA of HSV-1 encodes approximately 80 different proteins. However, the virus does not have its own machinery for<br />
expressing those genes; instead, the virus must divert the gene expression machinery of the host cell. That process is triggered<br />
by a viral regulatory protein designated VP16, whose function is to stimulate transcription of the first viral genes to be expressed<br />
in the infected cell (the immediate-early, or IE, genes).<br />
Chromatin-modifying coactivators in herpes virus infection and a paradox<br />
The strands of DNA in which the human genome is encoded are much longer than the diameter of a typical human cell. To help<br />
fit the DNA into the space of a cell, eukaryotic DNA is typically packaged as chromatin, in which the DNA is wrapped around<br />
“spools” of histone proteins, and these spools are then further arranged into higher-order structures. This elaborate packaging<br />
creates a problem when access is needed to the information carried in the DNA, such as when particular genes need to be<br />
expressed. This problem is solved in part by chromatin-modifying coactivator proteins, which either chemically change the<br />
histone proteins or else slide or remove them.<br />
Transcriptional activator proteins such as VP16 can recruit these chromatin-modifying coactivator proteins to target genes. We<br />
have shown this to be true for artificial reporter genes in human cells or in yeast, and it’s also true for the viral genes that VP16<br />
activates during an active infection. Curiously, however, the DNA of herpes simplex virus is not wrapped around histones inside<br />
the viral particle, and it also seems to stay free of histones inside the infected cell. That observation leads to a paradox: why<br />
would VP16 recruit chromatin-modifying coactivators to the viral DNA if the viral DNA doesn’t have any chromatin to modify?<br />
52
VARI | <strong>2009</strong><br />
We took several approaches to test whether the coactivators that are recruited to viral DNA by the VP16 activation domain<br />
really play a significant role in transcriptional activation. In some experiments, we knocked down expression of given coactivators<br />
using short interfering RNAs (siRNAs) and then measured viral gene expression during subsequent infection by HSV-1.<br />
In other experiments, we used cell lines that have mutations disrupting the expression or activity of a given coactivator. A<br />
third set of experiments used curcumin, derived from the curry spice turmeric, which is thought to inhibit the enzymatic<br />
activity of certain coactivators. In each of these situations, we expected to find that viral gene expression was inhibited, but<br />
the experiments yielded unexpected results: in each case, expression of the viral genes was essentially unaffected. We were<br />
forced to conclude that our initial hypothesis was wrong; the coactivators, although present, are not required for viral gene<br />
expression during lytic infection.<br />
The death of one hypothesis, however, gives life to new ideas. After the initial infection of a cold sore subsides, herpes simplex<br />
virus establishes a lifelong latent infection in sensory neurons. In the latent state, the viral genome is essentially quiet: very few<br />
viral genes are expressed. Moreover, the viral genome becomes packaged in chromatin much like the silent genes of the host<br />
cell. So our new hypothesis is that the coactivators recruited by VP16 are required to reactivate the viral genes from the latent<br />
or quiescent state. We’ve begun to test that hypothesis in quiescent infections in cultured cells, but the key tests will be in<br />
whole organisms with genuinely latent herpesvirus infections.<br />
Gene activation during cold acclimation of plants<br />
Although plants and their cells obviously have very different forms and functions than animals and their cells, the mechanisms used<br />
for expressing genetic information are quite similar. For the past decade, we have explored the role of chromatin-modifying coactivators<br />
in regulating genes that are turned on in low-temperature conditions. Some plants, including the prominent experimental<br />
organism Arabidopsis, can sense low (but nonfreezing) temperature in a way that provides protection from subsequent freezing<br />
temperatures, a process known as cold acclimation. We have collaborated with Michael Thomashow, a plant scientist at Michigan<br />
State University, to explore the mechanisms involved in activating genes during cold acclimation. To this point, we have focused<br />
on one particular histone acetyltransferase, termed GCN5, and two of its accessory proteins, ADA2a and ADA2b. Mutations in the<br />
genes encoding these coactivator proteins result in diminished expression of cold-regulated genes. Moreover, histones located<br />
at these cold-regulated genes become more highly acetylated during initial stages of cold acclimation. However, contrary to our<br />
expectations, the GCN5 and ADA2 proteins are not responsible for this cold-induced acetylation. In fact, we’ve tested several other<br />
Arabidopsis histone acetyltransferases, and none (on their own) seem solely responsible for this acetylation. It seems likely that<br />
redundant mechanisms are at work, such that when we disrupt one pathway, another pathway compensates.<br />
We also collaborated with groups in Greece and Pennsylvania to explore the distinct biological activities of the two ADA2<br />
proteins. Although the two proteins have very similar sequences and both are expressed throughout the plant, mutations in the<br />
genes encoding these two proteins have very different phenotypes. The ada2b mutants are very short, have smaller cells than<br />
normal, and are sterile. In contrast, the ada2a mutants seem quite normal in most attributes. Plants with mutations in both ADA2a<br />
and ADA2b are strikingly similar to plants with mutations in GCN5. We suspect that GCN5 can partner with either ADA2a or<br />
ADA2b and that these two distinct complexes affect different sets of genes and thus different developmental and stress response<br />
pathways. This work may help us understand whether the mechanisms by which plants express their genes can be modulated<br />
so as to protect crops from loss in yield or viability due to environmental stresses such as low temperature.<br />
53
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
External Collaborators<br />
Kanchan Pavangadkar and Michael F. Thomashow, Michigan State University, East Lansing<br />
Amy S. Hark, Muhlenberg College, Allentown, Pennsylvania<br />
Kostas Vlachonasios, Aristotle University of Thessaloniki, Greece<br />
Recent Publications<br />
Lu, Xu, and Steven J. Triezenberg. In press. Chromatin assembly on herpes simplex virus genomes during lytic<br />
infection. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms.<br />
Hark, Amy T., Konstantinos E. Vlachonasios, Kanchan A. Pavangadkar, Sumana Rao, Hillary Gordon, Ioannis-<br />
Dimosthenis Adamakis, Athanasios Kaldis, Michael F. Thomashow, and Steven J. Triezenberg. <strong>2009</strong>. Two Arabidopsis<br />
orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochimica et Biophysica Acta -<br />
Gene Regulatory Mechanisms 1789(2): 117–124.<br />
Kutluay, Sebla B., Sarah L. DeVos, Jennifer E. Klomp, and Steven J. Triezenberg. <strong>2009</strong>. Transcriptional coactivators<br />
are not required for herpes symplex virus type 1 immediate-early gene expression in vitro. Journal of Virology 83(8):<br />
3436–3449.<br />
Kutluay, Sebla B., and Steven J. Triezenberg. <strong>2009</strong>. Regulation of histone deposition on the herpes simplex virus type<br />
1 genome during lytic infection. Journal of Virology 83(11): 5835–5845.<br />
Kutluay, Sebla B., and Steven J. Triezenberg. <strong>2009</strong>. Role of chromatin during herpesvirus infections. Biochimica et<br />
Biophysica Acta 1790(6): 456–466.<br />
From left: Testori, Alberts, Klomp, Kutluay, Triezenberg, Rappley, Lu<br />
54
VARI | <strong>2009</strong><br />
George F. Vande Woude, Ph.D.<br />
Laboratory of Molecular Oncology<br />
Dr. Vande Woude received his M.S. (1962) and Ph.D. (1964) from Rutgers University. From 1964–<br />
1972, he served first as a postdoctoral research associate, then as a research virologist for the U.S.<br />
Department of Agriculture at Plum Island Animal Disease Center. In 1972, he joined the National<br />
Cancer Institute as Head of the Human Tumor Studies and Virus Tumor Biochemistry sections and,<br />
in 1980, was appointed Chief of the Laboratory of Molecular Oncology. In 1983, he became Director<br />
of the Advanced Bioscience Laboratories–Basic Research Program at the National Cancer Institute’s<br />
Frederick Cancer Research and Development Center, a position he held until 1998. From 1995, Dr.<br />
Vande Woude first served as Special Advisor to the Director, and then as Director for the Division of<br />
Basic Sciences at the National Cancer Institute. In 1999, he was recruited to become the founding<br />
Director of the Van Andel Research Institute. In <strong>2009</strong>, Vande Woude stepped down as Director and<br />
assumed the new title of Distinguished <strong>Scientific</strong> Fellow, while retaining his role of head of the Laboratory<br />
of Molecular Oncology.<br />
Staff<br />
Laboratory Staff<br />
Students<br />
Visiting Scientists<br />
Qian Xie, M.D., Ph.D.<br />
Yu-Wen Zhang, M.D., Ph.D.<br />
Chongfeng Gao, Ph.D.<br />
Carrie Graveel, Ph.D.<br />
Dafna Kaufman, M.Sc.<br />
Angelique Berens, B.S.<br />
Jack DeGroot, B.S.<br />
Curt Essenburg, B.S.<br />
Betsy Haak, B.S.<br />
Liang Kang, B.S.<br />
Rachel Kuznar, B.S.<br />
Benjamin Staal, B.S.<br />
Ryan Thompson, B.S.<br />
Yanli Su, A.M.A.T<br />
Laura Holman<br />
Amy Nelson<br />
Ala’a Abughoush<br />
Sara Kunz<br />
Kathleen Pollock<br />
David Wenkert, M.D.<br />
Yuehai Shen, Ph.D.<br />
Edwin Chen, B.S.<br />
55
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
The Laboratory of Molecular Oncology is focused on understanding the numerous and diverse roles that MET and HGF/SF<br />
play in malignant progression and metastasis. Our work involves a wide variety of cancers, animal models, and drug therapies.<br />
The combination of studies, coupled with our examination of MET signaling, will lead to a greater understanding of tumor<br />
progression and new knowledge for developing and delivering novel targeted therapies.<br />
Tumor phenotypic switching<br />
Malignant progression leading to metastasis is the primary cause of death due to cancer. Metastasis begins with proliferating<br />
tumor cells that become invasive and detach from the primary tumor mass, invading the extracellular matrix, entering the<br />
bloodstream or lymphatic vessels, and establishing metastases or secondary tumors as proliferating colonies at distant sites.<br />
Since phenotypic switching from proliferative to invasive and the return to proliferative is crucial for malignant progression, we<br />
use in vitro and in vivo methods to select proliferative and invasive subclones from tumor cell populations.<br />
We explored the signal pathway underlying phenotypic switching by integrative genomic studies including gene expression<br />
analysis, spectral karyotyping (SKY), and fluorescent in situ hybridization (FISH). We observed that subtle and specific changes<br />
in chromosome content ratio are virtually the same as the changes in the chromosome transcriptome ratio, showing that major<br />
changes in gene expression are mediated by gains or losses in chromosome content. Importantly, a significant number of the<br />
genes whose expression change is greater than twofold are functionally consistent with changes in the proliferative or invasive<br />
phenotypes. Our results imply that chromosome instability can provide the diversity of gene expression that allows a tumor to<br />
switch between proliferative and invasive phenotypes during tumor progression.<br />
Met in murine mammary tumors and human basal breast cancers<br />
Understanding the signaling pathways that drive aggressive breast cancers is critical to the development of effective<br />
therapeutics. The oncogene MET is associated with decreased survival in breast cancer, yet the role it plays in the various<br />
breast cancer subtypes is unclear. We are investigating the role that this oncogene plays in breast cancer progression and<br />
metastasis by using a novel mouse model of mutationally activated Met (Met mut ). We discovered that mutationally activated<br />
Met induces a high incidence of diverse mammary tumors in mice, and these Met mut mice tumors have several characteristics<br />
similar to those of aggressive human breast cancers, such as the absence of progesterone receptor and ERBB2 expression.<br />
These results led us to examine how MET is associated with the various human breast cancer subtypes. With gene<br />
expression and tissue microarray analysis, we observed that high MET expression in human breast cancers significantly<br />
correlated with estrogen receptor–negative/ERBB2-negative tumors and with basal breast cancers. Few treatment options<br />
exist for breast cancers of the basal or trastuzumab-resistant ERBB2 subtypes. We conclude from these studies that MET<br />
is a key oncogene in the development of the most aggressive breast cancer subtypes and may be a significant therapeutic<br />
target. Currently, we are investigating the similarities and differences in signaling pathways involved in MET-driven versus<br />
ERBB2-driven breast cancers.<br />
Tumor xenograft models for preclinical testing of MET drugs<br />
Aberrant activation of the HGF-Met signaling pathway is one of the causal events in cancer development and progression<br />
and is frequently observed in almost all types of human cancers. MET is becoming an ideal target for cancer intervention,<br />
and the movements toward developing MET drugs are very active. In the past several years, many drugs targeting the<br />
HGF-MET pathway have been developed, including neutralizing antibodies against HGF or MET and various small-molecule<br />
kinase inhibitors of MET. This has resulted in the need for suitable animal models for preclinically testing the drug efficacies<br />
in vivo.<br />
56
VARI | <strong>2009</strong><br />
We had previously generated a transgenic mouse that produces human HGF in the severe combined immune deficiency<br />
(SCID) background. This animal model provides species-compatible ligand for human MET and is ideal for investigating<br />
paracrine MET signaling in human cancer cells (mouse HGF has very low activity on human MET). We found that, compared<br />
with control SCID mice, the human HGFtg-SCID (huHGFtg-SCID) mice significantly enhance tumor xenograft growth of many<br />
MET-positive human cancer cells derived from lung, breast, stomach, colon, kidney, and pancreas. Currently, we are using<br />
those xenograft models established in the huHGFtg-SCID mice for testing MET drugs alone or in combination with other cancer<br />
drugs. Meanwhile, we are also developing metastatic models in the huHGFtg-SCID mice.<br />
In vivo modeling of glioblastoma multiforme<br />
Glioblastoma multiforme (GBM) is one of the most devastating cancers. The hallmark of GBM is the invasiveness of the<br />
tumor cells infiltrating into normal brain parenchyma, making it virtually impossible to remove the tumor completely by surgery<br />
and inevitably leading to recurrent disease. Progress in understanding GBM pathobiology and in developing novel antitumor<br />
therapies could be greatly accelerated with animal model systems that display characteristics of human GBM and that enable<br />
tumor monitoring through noninvasive imaging in real time. Subjecting human cancer cells to an experimental metastasis assay<br />
(ELM) often yields highly metastatic cells with higher proliferative and invasive potential. However, the ELM assay has not been<br />
tested previously with GBM, most likely because extracranial metastases of human GBM are clinically rare.<br />
In this study, we used ELM to enrich metastatic cell populations and found that three of four commonly used GBM lines<br />
(U251, U87, and DBTRG-05MG) were highly metastatic after repeated (M2) ELM selection. These GBM-M2 lines grew more<br />
aggressive orthotopically, and all showed significant multifold increases in IL6, IL8, MCP-1, and GM-CSF, which are cytokines<br />
and factors associated with poor GBM prognosis. DBM2 cells, derived from the DBTRG-05MG cell line, are highly invasive<br />
when grown as an orthotopic tumor (with areas of central necrosis, vascular hyperplasia, and intracranial dissemination),<br />
and also erode the skull, permitting the use of high-resolution micro-ultrasound in real time to non-invasively observe tumor<br />
growth and vascularization. We conclude that commonly used GBM cells have intrinsic metastatic potential which can be<br />
selected for in ELM assays. When implanted in the brain, the metastatic potential of GBM cells can be realized as a highly<br />
invasive phenotype. The DBM2 mouse model has characteristics that mimic the aggressively invasive behavior of clinical GBM,<br />
providing a valuable tool for investigating the factors that modulate glioblastoma growth, assessing invasion and vascularity,<br />
and evaluating novel therapeutic agents in real time. Currently we are in the process of using this model to test MET drugs<br />
and possible combinations for the purpose of blocking GBM invasion and studying the micro-environment of the host-tumor<br />
response to the treatment.<br />
The role of Mig-6 in cancer and joint disease<br />
The signaling mediated by receptor tyrosine kinases such as Met and EGFR plays a very important role in many developmental<br />
and physiological processes, and it is fine-tuned by many factors for proper action. Mitogen-inducible gene-6 (Mig-6), a<br />
scaffolding molecule, is one of the factors that can regulate Met and EGFR signaling through a negative feedback loop.<br />
Mig-6 is an immediate early response gene that can be rapidly up-regulated by growth factors like HGF and EGF, as well as<br />
by many stress stimuli such as mechanical stress. The Mig-6 gene locus is at human chromosome 1p36 that is frequently<br />
associated with various cancers. Studies in both humans and mice indicate that Mig-6 is a tumor suppressor gene. Decreased<br />
expression of Mig-6 is observed in several human cancers including breast, skin, pancreatic, and ovarian cancers, while<br />
targeted elimination of Mig-6 in mice leads to the development of neoplasms in the lung, gallbladder, bile duct, and skin. We<br />
also identified several Mig-6 gene mutations in lung cancer, even though mutation in Mig-6 seems to be a rare event. Besides<br />
its role in cancer, Mig-6 also plays an important role in maintaining normal joint function: its deficiency in mice results in the<br />
development of early-onset degenerative joint disease. Currently, we are investigating what roles Mig-6 may play in cancer<br />
development and in maintenance of joint function, and the mechanism of how losing Mig-6 activity leads to the pathological<br />
conditions of cancer and joint disease.<br />
57
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
External Research Collaborators<br />
Donald Bottaro and Benedetta Peruzzi, National Cancer Institute, Bethesda, Maryland<br />
Sandra Cottingham, Spectrum Health Hospital, Grand Rapids, Michigan<br />
Sherri Davies and Matthew Ellis, Washington University, St. Louis, Missouri<br />
Francesco DeMayo, Baylor College of Medicine, Houston, Texas<br />
Ermanno Gherardi, MRC Center, Cambridge, England<br />
Beatrice Knudsen, Fred Hutchinson Cancer Research Center, Seattle, Washington<br />
Stephanie Kermorgant, Queen Mary and Westfield College, University of London, U.K.<br />
Ernest Lengyel and Ravi Salgia, University of Chicago, Illinois<br />
Kangda Liu, Zhongshan Hospital, Fudan University, P.R.C.<br />
Patricia LoRusso and Fred Miller, Barbara Ann Karmanos Cancer Institute, Detroit, Michigan<br />
Benjamin Neel, University of Toronto, Ontario, Canada<br />
Ilan Tsarfaty, Tel Aviv University, Israel<br />
Robert Wondergem, East Tennessee State University, Johnson City<br />
Recent Publications<br />
Gao, C.-F., Q. Xie, Y.-W. Zhang, Y. Su, P. Zhao, B. Cao, K. Furge, J. Sun, K. Rex, T. Osgood, et al. <strong>2009</strong>. Therapeutic<br />
potential of hepatocyte growth factor/scatter factor neutralizing antibodies: inhibition of tumor growth in both authcrine and<br />
paracrine hepatocyte growth factor/scatter factor:c-Met–driven models of leiomyosarcoma. Molecular Cancer Therapeutics<br />
8(10): 2803–2810.<br />
Graveel, C.R., J.D. DeGroot, Y. Su, J.M. Koeman, K. Dykema, S. Leung, J. Snider, S.R. Davies, P.J. Swiatek,<br />
S. Cottingham, et al. <strong>2009</strong>. Met induces diverse breast carcinomas in mice and is associated with human basal breast cancer.<br />
Proceedings of the National Academy of Sciences U.S.A. 106(31): 12909–12914.<br />
Kort, Eric J., Nigel Paneth, and George F. Vande Woude. <strong>2009</strong>. The decline in U.S. cancer mortality in people born since<br />
1925. Cancer Research 69(16): 6500–6505.<br />
VanBrocklin, Matthew W., James P. Robinson, Todd Whitwam, Adam R. Guibeault, Julie Koeman, Pamela J. Swiatek,<br />
George F. Vande Woude, Joseph D. Khoury, and Sheri L. Holmen. <strong>2009</strong>. Met amplification and tumor progression in<br />
Cdkn2a-deficient melanocytes. Pigment Cell & Melanoma Research 22(4): 454–460.<br />
Standing, from left: Zhang, Nelson, Essenburg, Graveel, Staal, Su, DeGroot, Thompson, Haak, Xie, Gao;<br />
seated, from left: Holman, Vande Woude, Kaufman<br />
58
VARI | <strong>2009</strong><br />
Craig P. Webb, Ph.D.<br />
Program for Translational Medicine<br />
Dr. Webb received his Ph.D. in cell biology from the University of East Anglia, England, in 1995. He<br />
then served as a postdoctoral fellow in the laboratory of George Vande Woude in the Molecular<br />
Oncology Section of the Advanced BioScience Laboratories–Basic Research Program at the National<br />
Cancer Institute, Frederick Cancer Research and Development Center, Maryland (1995–1999). Dr.<br />
Webb joined VARI as a <strong>Scientific</strong> Investigator in October 1999; he now oversees the Program for<br />
Translational Medicine as Senior <strong>Scientific</strong> Investigator.<br />
Staff<br />
Students<br />
Visiting Scentist<br />
David Cherba, Ph.D.<br />
Jessica Hessler, Ph.D.<br />
Jeremy Miller, Ph.D.<br />
David Monsma, Ph.D.<br />
Emily Eugster, M.S.<br />
Patrick Richardson, M.S.<br />
Sujata Srikanth, M.Phil.<br />
Dawna Dylewski, B.S.<br />
Brian Hillary, B.A.<br />
Hailey Jahn, B.S.<br />
Marcy Ross, B.S.<br />
Stephanie Scott, B.S.<br />
Theresa Wood, B.A.<br />
Katherine Koehler<br />
Nicole Beuschel<br />
Orrie Close<br />
Bess Connors<br />
Molly Dobb<br />
Phillip Dumas<br />
Sean Vance<br />
Richard Leach, M.D.<br />
59
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Research Interests<br />
Molecular biomarkers are widely expected to revolutionize the current trial-and-error practice of medicine by enabling a<br />
more predictive discipline in which therapies are targeted toward the molecular constitution of individual patients and their<br />
disease. This concept is often termed “personalized medicine”. Biomarkers are being widely evaluated for their ability to<br />
assess disease risk, detect and monitor disease over time, accurately identify disease stage, approximate prognosis, and<br />
predict optimal targeted treatments. The Program for Translational Medicine was launched in 2006 to extend the Institute’s<br />
translational research capabilities, with a focus on the development of molecular biomarker strategies with clinical implications.<br />
The program’s activities have focused on building the critical translational infrastructure and technologies, fostering clinical and<br />
industrial partnerships, and coordinating the multidisciplinary project teams required to implement molecular-based approaches<br />
in medicine. The Program of Translational Medicine, with its multidisciplinary partners, strives to create an efficient pipeline<br />
between the clinic and the research laboratory for efficient discovery and clinical application of novel biomarker strategies. We<br />
also work to increase the readiness of the community to implement advances in molecular medicine, benefiting human health<br />
and promoting West Michigan as a leader in biomarker research.<br />
Translational informatics<br />
To accelerate the implementation of personalized medicine, the consolidation and real-time analysis of standardized molecular<br />
and clinical/preclinical data is critical. Thus, much of our effort over the past several years has focused on the development of<br />
an integrated informatics solution known as the XenoBase BioIntegration Suite (XB-BIS; see http://xbtransmed.com). XB-BIS<br />
supports essential features of data management, data analysis, knowledge management, and reporting within an integrated<br />
framework, enabling the efficient exchange of information between the basic research laboratory and the clinic. XB-BIS has<br />
recently been licensed to industrial and academic partners with an interest in biomarker research and the development of<br />
molecular-based diagnostics; these include Children’s Memorial Medical Center, Qiagen, and Sequenom.<br />
Community partnerships and economic development<br />
Productive partnerships are pivotal to our efforts in biomarker research and personalized medicine. In the Center for Molecular<br />
Medicine, the Van Andel Institute and Spectrum Health Hospitals created a CLIA-certified/CAP-accredited clinical diagnostics<br />
laboratory for biomarker qualification and the development of associated diagnostic assays. This entity was recently acquired<br />
by Sequenom, but it continues to offer cutting-edge molecular diagnostic tests and remains central to our personalized<br />
medicine initiatives. For example, ongoing research activities with Sequenom are geared toward implementation of novel,<br />
molecular-based technologies into our personalized medicine initiative.<br />
ClinXus (http://www.clinxus.org) was developed to coordinate the emerging West Michigan translational research enterprise.<br />
In September 2006, ClinXus was awarded a Michigan 21st Century Jobs Fund grant to support early-stage development and<br />
operations, and it has membership in the Predictive Safety and Testing Consortium (PSTC) of the Critical Path Institute. The<br />
PSTC brings pharmaceutical companies together to share and validate each other’s safety testing methods under advisement<br />
of the FDA and the European Medicines Agency. Membership in this consortium will help ensure that West Michigan remains<br />
at the forefront of biomarker research and development and will further the community’s rapidly emerging life sciences and<br />
health care industry.<br />
60
VARI | <strong>2009</strong><br />
Predictive therapeutics protocol<br />
Translational research represents the interface where hypotheses advance to studies that ultimately provide definitive information<br />
for a clinical decision. A fundamental challenge in clinical cancer research remains how to make best use of current<br />
biomarker technologies, advances in computational biology, the expanding pharmacopeia, and a rapidly expanding knowledge<br />
of disease networks to deliver targeted treatments to cancer patients with optimal therapeutic index. Our research is focused<br />
on developing, testing, and refining biomarker-driven analytical methods to systematically predict combinations of drugs that<br />
target the tumor. We are also considering the means by which such information should be conveyed to the treating physician<br />
in support of medical decision-making.<br />
In 2008, we completed a feasibility study of 50 late-stage pediatric and adult cancer patients in which tumor-derived gene<br />
expression profiles were analyzed to identify potential drugs to target perturbed molecular components of each patient’s<br />
specific tumor. With patient consent, tumor biopsies were collected, qualified by pathology, and processed within the CMM to<br />
generate a standardized gene expression profile for the tumor. These molecular data were uploaded into XB-BIS along with<br />
pertinent clinical data and compared with other patient samples within the database. Within XB-BIS, deregulated patterns<br />
of gene expression were identified and analyzed to identify drugs that have predicted efficacy based upon their molecular<br />
mechanism of action and the tumor’s genomic data. A report scoring a series of drugs for predicted efficacy was generated<br />
within XB-BIS and conveyed to the treating physician in an actionable, electronic format for consideration in treatment planning.<br />
To be compatible with real-time prospective decision making, the process from patient consent to molecular report had to be<br />
completed in 5-10 days. In parallel, a series of tumor grafts was established in immune-compromised mice, which closely<br />
resembles the human disease at the phenotypic and molecular level. This resource is currently being used to test biomarkerdriven<br />
predictive models (and the identified drugs) in a more systematic fashion and to evaluate novel targeted agents. Over the<br />
long term, the treatments are captured within XB-BIS together with critical outcome variables, allowing the predictive analytical<br />
methods to be refined and optimized.<br />
Anecdotal signs of success in a handful of patients have provided the impetus to launch a series of follow-up studies with an<br />
expanded patient population using a more rigorous statistical design. Collaborative studies on glioblastoma through The Ben and<br />
Catherine Ivy Foundation and on neuroblastoma through the Vermont Cancer Center and University of Vermont are planned for<br />
<strong>2009</strong>. In conjunction with our laboratory efforts to isolate, characterize, and target the putative cancer stem-cell subpopulation of<br />
metastatic tumors, biomarker-driven approaches that identify a rational treatment regimen targeting the molecular composition<br />
of the patient’s tumor hold promise for the future treatment of metastatic and refractory malignancies.<br />
From left: Jahn, Srikanth, Richardson, Koehler, Monsma, Dylewski, Eugster, Scott, Miller, Wood, Dumas,<br />
Cherba, Webb<br />
61
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
External Collaborators<br />
Academic Surgical Associates, Advanced Radiology Services, Cancer & Hematology Centers of Western Michigan, P.C.,<br />
DeVos Children’s Hospital, Digestive Disease Institute, Grand Valley Medical Specialists, Grand Valley State University,<br />
MMPC, Saint Mary’s Health Care, Spectrum Health, and West Michigan Heart, all of Grand Rapids, Michigan<br />
Barbara Ann Karmanos Institute and Henry Ford Hospital, Detroit, Michigan<br />
The Ben and Catherine Ivy Foundation, Palo Alto, California<br />
Case Western Reserve University School of Medicine, Cleveland, Ohio<br />
GeneGo, Inc., and Oncology Care Associates, St. Joseph, Michigan<br />
Jackson Laboratory-West, Sacramento, California<br />
Jasper Clinical Research & Development, Inc., Kalamazoo, Michigan<br />
Johns Hopkins University, Baltimore, Maryland<br />
M.D. Anderson Cancer Center, Houston, Texas<br />
Mary Crowley Cancer Center, Dallas, Texas<br />
Mayo Clinic, Rochester, Minnesota<br />
Michigan State University, East Lansing<br />
New York University, New York City<br />
The Ohio State University, Columbus<br />
Pfizer, Ann Arbor, Michigan; Saint Louis, Missouri; Groton, Connecticut<br />
TGen, Phoenix, Arizona<br />
University of Alabama at Birmingham<br />
University of California, San Francisco<br />
University of Michigan, Ann Arbor<br />
Vermont Cancer Center and University of Vermont<br />
Recent Publications<br />
Littman, B., J. Thompson, and C.P. Webb. In press. Where are we heading/What do we really need? In Biomarkers in Drug<br />
Development: A Handbook of Practice, Application, and Strategy, Michael Bleavins, Ramin Rahbari, Malle Jurima-Romet, and<br />
Claudio Carini, eds. New York; Wiley.<br />
Ivanov, S.V., J. Miller, R. Lucito, C. Tang, A.V. Ivanova, J. Pei, M. Carbone, C. Cruz, A. Beck, C. Webb, et al. <strong>2009</strong>. Genomic<br />
events associated with progression of pleural malignant mesothelioma. International Journal of Cancer 124(3): 589–599.<br />
Webb, C.P., and D. Cherba. <strong>2009</strong>. Systems biology of personalized medicine. In Bioinformatics for Systems Biology:<br />
Second Edition, Introduction to Informatics, Stephen Krawetz, ed. New York: Humana, pp. 615–630.<br />
62
VARI | <strong>2009</strong><br />
Retina whole mount<br />
Images of mouse retina whole mounts, postnatal day 14. Red stain is GSA lectin on endothelial blood vessel cells; green stain is FITC dextran<br />
perfusion of blood vessels. At the right center of the red image is a clump of remnant hyaloid blood vessels; few of these vessels can be seen in<br />
the green image because they no longer have blood flow. Original magnification 40×. Photo by Jennifer Bromberg-White of the Duesbery lab.<br />
63
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Michael Weinreich, Ph.D.<br />
Laboratory of Chromosome Replication<br />
Dr. Weinreich received his Ph.D. in biochemistry from the University of Wisconsin–Madison in 1993. He<br />
then was a postdoctoral fellow in the laboratory of Bruce Stillman, Director of the Cold Spring Harbor<br />
Laboratory, New York, from 1993 to 2000. Dr. Weinreich joined VARI as a <strong>Scientific</strong> Investigator in<br />
March 2000. He was promoted to Senior <strong>Scientific</strong> Investigator in September 2008.<br />
Staff<br />
Dorine Savreux, Ph.D.<br />
FuJung Chang, M.S.<br />
Carrie Gabrielse, B.S.<br />
Students<br />
Ying-Chou Chen, M.S.<br />
Charles Miller, B.S.<br />
Christina Gourlay<br />
Caitlin May<br />
Christina Untersperger<br />
64
VARI | <strong>2009</strong><br />
Research Interests<br />
We study early events that promote the initiation of DNA synthesis, which occurs at specific sequences termed replication<br />
origins. Various genome-wide approaches have identified from 320 to 420 possible replication origins in budding yeast, and<br />
there are perhaps 10,000 origins in human cells. The initiation of DNA replication occurs in a temporally distinct manner during<br />
G1 and S phase, and no origin initiates replication (fires) more than once per cell cycle to maintain normal diploid content. In<br />
G1 phase, each origin assembles approximately 40 polypeptides in a temporally defined order, culminating in the initiation of<br />
DNA replication at the G1/S phase boundary. The first stage of this process is called pre-replicative complex assembly and<br />
requires the origin recognition complex (ORC), Cdc6, and Cdt1. ORC directly binds to origin sequences and then recruits Cdt1<br />
and Cdc6 during G1 phase. These three proteins cooperate to load the MCM DNA helicase at origins in an ATP-dependent<br />
reaction. Cyclin-dependent kinases and the Cdc7-Dbf4 kinase then catalyze the association of additional proteins with the<br />
MCM helicase to activate it, ultimately causing unwinding of the duplex DNA and the initiation of bidirectional DNA synthesis<br />
(Figure 1). In our lab we study three key aspects of DNA replication:<br />
1) Replication origin structure<br />
2) How Cdc6-ATP functions to load the MCM helicase within a chromatin context<br />
3) How Cdc7-Dbf4 kinase contributes to the normal cell cycle and human malignancies<br />
Figure 1<br />
Studies to understand the basic molecular biology of DNA replication and cell cycle progression are highly relevant for cancer<br />
biology, given that malignant cells often contain mutations in the cell growth and checkpoint pathways that drive normal<br />
proliferation. Here we describe recent studies on the Cdc7-Dbf4 kinase, which is a crucial regulator of DNA replication in all<br />
eukaryotic cells.<br />
Cdc7-Dbf4 is a conserved, two-subunit, serine/threonine protein kinase that catalyzes DNA synthesis at individual replication<br />
origins. Cdc7-Dbf4 promotes DNA synthesis after MCM helicase loading at the origin, likely by activating its helicase activity.<br />
This leads to origin unwinding and the assembly of DNA polymerases that initiate bidirectional DNA synthesis. Although Cdc7<br />
is a member of the protein kinase superfamily, it requires the Dbf4 regulatory subunit to activate its kinase activity. We have<br />
determined the regions of Dbf4 that bind to and activate Cdc7 kinase by mutational analysis, and we are also investigating how<br />
Dbf4 targets Cdc7 kinase to its various substrates in the cell.<br />
65
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Figure 2<br />
Figure 2. Functional regions of Dbf4 determined by deletion analysis. Dbf4 contains three regions called motifs N, M, and C that are<br />
conserved among Dbf4 orthologs. A BRCT-like domain spans motif N. Motif C encodes a single C 2<br />
H 2<br />
type Zn-finger. Finally, two regions<br />
spanning motif M and C interact with Cdc7 and activate its kinase activity.<br />
In Figure 2, we summarize our analysis of Dbf4 functional regions. The N-terminal third of Dbf4 is dispensable for DNA replication.<br />
However, the N-terminus encodes several functions that are important for Dbf4 function. There are two putative classic<br />
nuclear localization sequences (NLSs) at 55-61 and 251-257. Although the first sequence is dispensable, deletion of both<br />
sequences is lethal. Functionality is restored to a dbf4 mutant lacking the first 265 residues by addition of a heterologous NLS<br />
from the SV40 large T-antigen, suggesting that there are no NLS sequences in the Dbf4 C-terminus.<br />
We also identified a BRCT-like domain from residues 115-219 that is apparently conserved in all Dbf4 orthologs. BRCT<br />
domains are often present in DNA damage-responsive proteins and they interact with phosphorylated residues. Although Dbf4<br />
mutants deleting this region are viable, they exhibit a slow S-phase and defects in response to DNA-damaging agents such as<br />
hydroxyurea, bleomycin, and methylmethane sulfonate. Whether the BRCT-like region governs a DNA repair function for Dbf4<br />
is uncertain, because addition of an SV40 NLS to the dbf4-ND221 deletion mutant reverses most of these DNA replication and<br />
damage phenotypes. This suggests that the damage sensitivity is a secondary consequence of lowered nuclear localization<br />
and, therefore, compromised initiation activity. Consistent with this explanation, we found that many initiation mutants also<br />
exhibit secondary DNA damage sensitivities. Interestingly, deletion of the BRCT-like domain causes defects in late-origin<br />
activation, but early origins are activated normally. This raises the intriguing possibility that the BRCT-like domain targets the<br />
kinase to late replication origins.<br />
We also constructed a series of C-terminal deletion Dbf4 mutants; such mutants that remove a conserved Zn-finger motif are<br />
viable. This indicates that C-terminal residues are not essential for Dbf4 activity. However, deletion of C-terminal residues<br />
results in a markedly slower S-phase progression, temperature sensitivity, and DNA damage sensitivity. This suggests that the<br />
C-terminus is required to activate full Cdc7 kinase activity or to target it to important replication substrates. Using recombinant<br />
Cdc7 and Dbf4 proteins, we found that Dbf4 mutants lacking the C-terminus have a profound defect in Cdc7 kinase activation.<br />
The Zn-finger motif also interacts with Cdc7 via a two-hybrid assay, and this interaction depends on conserved residues in the<br />
Zn-finger. Lastly, there is a second Cdc7 binding site that overlaps motif M. We found that either Cdc7 binding region could<br />
be deleted individually and still allow Cdc7 binding, but deletion of both domains does not allow Cdc7 binding.<br />
We would like to identify proteins that interact with the Dbf4 N-terminus and determine the functional consequences of those<br />
interactions. Clearly the N-terminal third of Dbf4 is not required for DNA replication, but these residues are conserved in mouse<br />
and human cells and so must confer some critical function. Using a two-hybrid approach, we found that the Dbf4 N-terminus<br />
interacts with Polo kinase, a key regulator of mitotic progression. Detailed analysis of this interaction suggests that Dbf4<br />
influences chromosome segregation, which represents a totally new activity for Cdc7-Dbf4 kinase.<br />
66
VARI | <strong>2009</strong><br />
External Collaborators<br />
Catherine Fox, University of Wisconsin–Madison<br />
Carol Newlon, University of Medicine and Dentistry of New Jersey, Newark<br />
Philippe Pasero, CNRS, Montpellier, France<br />
Alain Verreault, University of Montreal, Quebec, Canada<br />
Wolfgang Zachariae, Max Plank Institute, Dresden, Germany<br />
Recent Publications<br />
Harkins, V., Carrie Gabrielse, L. Haste, and M. Weinreich. In press. Budding yeast Dbf4 sequences required for Cdc7 kinase<br />
activation and identification of a functional relationship between the Dbf4 and Rev1 BRCT domains. Genetics.<br />
Miller, Charles T., Carrie Gabrielse, Ying-Chou Chen, and Michael Weinreich. <strong>2009</strong>. Cdc7p-Dbf4p regulates mitotic exit by<br />
inhibiting polo kinase. PLoS Genetics 5(5): e1000498.<br />
Bonte, Dorine, Charlotta Lindvall, Hongyu Liu, Karl Dykema, Kyle Furge, and Michael Weinreich. 2008. Cdc7-Dbf4 kinase<br />
overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia 10(9): 920–931.<br />
Chang, FuJung, James F. Theis, Jeremy Miller, Conrad A. Nieduszynski, Carol S. Newlon, and Michael Weinreich. 2008.<br />
Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW<br />
sequence within the origin recognition complex binding site. Molecular and Cellular Biology 28(16): 5071–5081.<br />
Fox, Catherine A., and Michael Weinreich. 2008. Beyond heterochromatin: SIR2 inhibits the initiation of DNA replication.<br />
Cell Cycle 7(21): 3330–3334.<br />
From left: Savreux, Gabrielse, Chang, Miller, Chen, Weinreich<br />
67
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Bart O. Williams, Ph.D.<br />
Laboratory of Cell Signaling and Carcinogenesis<br />
Dr. Williams received his B.S. degree from Carroll College in Waukesha, Wisconsin. In 1996, he received<br />
his Ph.D. in Biology from the Massachusetts Institute of Technology for studies in the laboratory<br />
of Tyler Jacks characterizing mouse models carrying mutations in the Rb and p53 genes. From 1996<br />
to 1999, he was a postdoctoral fellow at the National Institutes of Health in the laboratory of Harold<br />
Varmus, former Director of NIH. Dr. Williams joined VARI as a <strong>Scientific</strong> Investigator in July 1999 and<br />
was promoted to Senior <strong>Scientific</strong> Investigator in 2006.<br />
Staff<br />
Charlotta Lindvall, M.D., Ph.D.<br />
Alex Zhen-Dong Zhong, Ph.D.<br />
Cassandra Zylstra Diegel, B.S.<br />
Angela Lake, B.S.<br />
Ammar Saladhar, B.S.<br />
Kyle VanKoevering, B.S.<br />
Students<br />
Stephanie Berry<br />
Sathyanarayanan Elumalai<br />
Audrey Sanders<br />
Cassie Schumacher<br />
68
VARI | <strong>2009</strong><br />
Research Interests<br />
Our laboratory is interested in understanding how alterations in the Wnt signaling pathway cause human disease. Specifically,<br />
we have focused our efforts on the functions of the Wnt co-receptors, Lrp5 and Lrp6. Wnt signaling is an evolutionarily<br />
conserved process that functions in the differentiation of most tissues within the body. Given its central role in growth and<br />
differentiation, it is not surprising that alterations in the pathway are among the most common events associated with human<br />
cancer. In addition, several other human diseases, including osteoporosis, cardiovascular disease, and diabetes, have been<br />
linked to altered regulation of this pathway.<br />
A specific focus of work in our laboratory is characterizing the role of Wnt signaling in bone formation. Our interest is not only<br />
from the perspective of normal bone development, but also in trying to understand whether aberrant Wnt signaling plays a role<br />
in the predisposition of some common tumor types (for example, prostate, breast, lung, and renal tumors) to metastasize to<br />
and grow in bone. The long-term goal of this work is to provide insights useful in developing strategies to lessen the morbidity<br />
and mortality associated with skeletal metastasis.<br />
Wnt signaling in normal bone development<br />
Mutations in the Wnt receptor Lrp5 have been causally linked to alterations in human bone development. We have characterized<br />
a mouse strain deficient in Lrp5 and shown that it recapitulates the low-bone-density phenotype seen in human patients<br />
who have Lrp5 deficiency. We have further shown that mice carrying mutations in both Lrp5 and the related Lrp6 protein have<br />
even more-severe defects in bone density.<br />
To test whether Lrp5 deficiency causes changes in bone density due to aberrant signaling through b-catenin, we created mice<br />
carrying an osteoblast-specific deletion of b-catenin (OC-cre;b-catenin-flox/flox mice). In collaboration with Tom Clemens of<br />
the University of Alabama at Birmingham, we found that alterations of Wnt/b-catenin signaling in osteoblasts lead to changes<br />
in the expression of RANKL and osteoprotegerin (OPG). Consistent with this, histomorphometric evaluation of bone in the mice<br />
with osteoblast-specific deletions of either Apc or b-catenin revealed significant alterations in osteoclastogenesis.<br />
We are addressing how other genetic alterations linked to Wnt/b-catenin signaling affect bone development and osteoblast<br />
function. We have generated mice with conditional alleles of Lrp6 and Lrp5 that can be inactivated via cre-mediated recombination,<br />
and we are assessing the roles of these genes at different stages of osteoblast differentiation using both OC-cre and<br />
Dermo1-cre. Finally, we are working to determine what other signaling pathways may impinge on b-catenin signaling to control<br />
osteoblast differentiation and function.<br />
Wnt signaling in mammary development and cancer<br />
We are also addressing the relative roles of Lrp5 and Lrp6 in Wnt1-induced mammary carcinogenesis. A deficiency in Lrp5<br />
dramatically inhibits the development of mammary tumors, and a germline deficiency for Lrp5 or Lrp6 results in delayed<br />
mammary development. Because Lrp5-deficient mice are viable and fertile, we have focused our initial efforts on these mice.<br />
In collaboration with Caroline Alexander’s laboratory, we have found dramatic reductions in the number of mammary progenitor<br />
cells in these mice, and we are examining the mechanisms underlying this reduction. We have also found that Lrp6 plays a key<br />
role in mammary development, and we are focusing on the mechanisms underlying this unique role. Finally, we are defining<br />
the relative roles of b-catenin and mTOR signaling in the initiation and progression of Wnt1-induced mammary tumors. We are<br />
particularly interested in the role(s) of these pathways in regulating the proliferation of normal mammary progenitor cells, as well<br />
as of tumor-initiating cells.<br />
Wnt signaling in metabolic syndrome<br />
Several studies have linked mutations in Lrp5 and/or Lrp6 to the development of diabetes, dyslipedemias, and hypertension in<br />
humans and mice. We are exploring the roles of these genes in this context by creating mice carrying conditional deletions in<br />
hepatocytes or in adipocytes and evaluating their phenotypes.<br />
69
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Wnt signaling in prostate development and cancer<br />
Two hallmarks of advanced prostate cancer are the development of skeletal osteoblastic metastasis and the ability of the tumor<br />
cells to become independent of androgen for survival. The association of Wnt signaling with bone growth, plus the fact that<br />
b-catenin can bind to the androgen receptor and make it more susceptible to activation with steroid hormones other than DHT,<br />
make Wnt signaling an attractive candidate for explaining some phenotypes associated with advanced prostate cancer. We<br />
have created mice with a prostate-specific deletion of the Apc gene. These mice develop fully penetrant prostate hyperplasia<br />
by four months of age, and these tumors progress to frank carcinomas by seven months. We have found that these tumors<br />
initially regress under androgen ablation but show signs of androgen-independent growth some months later.<br />
VARI mutant mouse repository<br />
With support from the Institute, our laboratory maintains a repository of mutant mouse strains to support the general development<br />
of animal models of human disease.<br />
External Collaborators<br />
Bone development<br />
Mary Bouxsein, Beth Israel Deaconness Medical Center, Boston, Massachusetts<br />
Thomas Clemens, University of Alabama–Birmingham<br />
Marie-Claude Faugere, University of Kentucky, Lexington<br />
Fanxin Long and David Ornitz, Washington University, St. Louis, Missouri<br />
Merry Jo Oursler, Mayo Clinic, Rochester, Minnesota<br />
Matthew Warman, Harvard Medical School, Boston, Massachusetts<br />
Prostate cancer<br />
Wade Bushman, University of Wisconsin–Madison<br />
Valeri Vashioukhin, Fred Hutchinson Cancer Research Center, Seattle, Washington<br />
Mammary development<br />
Caroline Alexander, University of Wisconsin–Madison<br />
Yi Li, Baylor Breast Center, Houston, Texas<br />
Metabolic syndrome<br />
Tim Garvey, University of Alabama-Birmingham<br />
Jiandie Lin and Ormond MacDougald, University of Michigan, Ann Arbor<br />
Other organ systems/mechanisms of Wnt signaling<br />
Kathleen Cho and Eric Fearon, University of Michigan, Ann Arbor<br />
Kang-Yell Choi, Yansei University, Seoul, South Korea<br />
Silvio Gutkind, National Institute of Dental and Craniofacial Research, Bethesda, Maryland<br />
Kun-Liang Guan, University of California, San Diego<br />
Richard Lang and Aaron Zorn, Cincinnati Children’s Hospital Medical Center, Ohio<br />
Malathy Shekhar, Wayne State University, Detroit, Michigan<br />
70
VARI | <strong>2009</strong><br />
Recent Publications<br />
Li, Y., A. Ferris, B.C. Lewis, S. Orsulic, B.O. Williams, E.C. Holland, and S.H. Hughes. In press. The RCAS/TVA somatic<br />
gene transfer method in modeling human cancer. In Mouse Models for Cancer Research, J. Green and T. Ried, eds.,<br />
Springer Verlag.<br />
Williams, B.O., and K.L. Insogna. <strong>2009</strong>. Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling. Journal of Bone<br />
and Mineral Research 24(2): 171–178.<br />
Badders, Nisha M., Shruti Goel, Rod J. Clark, Kristine S. Klos, Soyoung Kim, Anna Bafico, Charlotta Lindvall,<br />
Bart O. Williams, and Caroline M. Alexander. <strong>2009</strong>. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells<br />
and is required to maintain the basal lineage. PLoS One 4(8): e6594.<br />
Castilho, Rogerio M., Cristiane H. Squarize, Lewis A. Chodosh, Bart O. Williams, and J. Silvio Gutkind. <strong>2009</strong>. mTOR<br />
mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 5(3): 279–289.<br />
Lindvall, Charlotta, Cassandra R. Zylstra, Nicole Evans, Richard A. West, Karl Dykema, Kyle A. Furge, and Bart O. Williams.<br />
<strong>2009</strong>. The Wnt co-receptor Lrp6 is required for normal mouse mammary gland development. PLoS One 4(6): e5813.<br />
Chen, Jindong, Kunihiko Futami, David Petillo, Jun Peng, PengFei Wang, Jared Knol, Yan Li, Sok Kean Khoo, Dan Huang,<br />
Chao-Nan Qian, et al. 2008. Deficiency of FLCN in mouse kidney led to development of polycystic kidneys and renal<br />
neoplasia. PLoS One 3(10): e3581.<br />
VanKoevering, K.K., and B.O. Williams. 2008. Transgenic mouse strains for conditional gene deletion during skeletal<br />
development. IBMS BoneKEy 5(5): 151–170.<br />
Zylstra, C.R., C. Wan, K.K. VanKoevering, A.K. Sanders, C. Lindvall, T.L. Clemens, and B.O Williams. 2008. Gene<br />
targeting approaches in mice: assessing the roles of LRP5 and LRP6 in osteoblasts. Journal of Muskuloskeletal<br />
& Neuronal Interactions 8(4): 291–293.<br />
Standing, from left: Zhong, Sanders, Lindvall, Lake, Elumalai;<br />
seated, from left: Diegel, Williams<br />
71
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
H. Eric Xu, Ph.D.<br />
Laboratory of Structural Sciences<br />
Dr. Xu went to Duke University and the University of Texas Southwestern Medical Center, where he<br />
earned his Ph.D. in molecular biology and biochemistry. Following a postdoctoral fellowship with Carl<br />
Pabo at MIT, he moved to GlaxoWellcome in 1996 as a research investigator in nuclear receptor drug<br />
discovery. Dr. Xu joined VARI as a Senior <strong>Scientific</strong> Investigator in July 2002 and was promoted to<br />
Distinguished <strong>Scientific</strong> Investigator in March 2007.<br />
Staff<br />
Students<br />
Visiting Scientists<br />
Abhishek Bandyopadhyay, Ph.D.<br />
Ajian He, Ph.D.<br />
Jiyuan Ke, Ph.D.<br />
Schoen Kruse, Ph.D.<br />
Raghu Malapaka, Ph.D.<br />
Karsten Melcher, Ph.D.<br />
Augie Pioszak, Ph.D.<br />
David Tolbert, Ph.D.<br />
Yong Xu, Ph.D.<br />
Chenghai Zhang, Ph.D.<br />
X. Edward Zhou, Ph.D.<br />
Jennifer Holtrop, B.S.<br />
Amanda Kovach, B.S.<br />
Naomi Parker, B.S.<br />
Kelly Powell, B.S.<br />
Debra Guthrey<br />
Cee Wah Chen<br />
Aoife Conneely<br />
Xiang Gao<br />
Clara Jurecky<br />
Shiva Kumar<br />
Kuntal Pal<br />
Emily Popma<br />
Leonor Ruivo<br />
Rachel Talaski<br />
Xiaoyong Zhi<br />
Jun Li, Ph.D.<br />
Ross Reynolds, Ph.D.<br />
72
VARI | <strong>2009</strong><br />
Research Interests<br />
My major research interest is the structures and functions of protein/ligand complexes that play key roles in major hormone<br />
signaling pathways. My secondary research interest is to explore the structural information with a goal of developing therapeutic<br />
agents for treating human disease, including cancer and diabetes. Research in my group currently focuses on three areas—<br />
nuclear hormone receptors, the Met tyrosine kinase receptor, and G protein–coupled receptors—because these proteins,<br />
beyond their fundamental roles in biology, are important drug targets. Our studies use multidisciplinary approaches, including<br />
molecular and cellular biology, biochemistry, animal physiology, and X-ray crystallography.<br />
Nuclear hormone receptors<br />
Nuclear hormone receptors form a large family comprising ligand-regulated and DNA-binding transcriptional factors, including<br />
receptors for classic steroid hormones such as estrogen, progesterone, androgens, and glucocorticoids, as well as receptors for<br />
peroxisome proliferator activators, vitamin D, vitamin A, and thyroid hormones. These classic receptors are among the most successful<br />
targets in the history of drug discovery: every receptor has one or more synthetic ligands currently being used as medicines.<br />
In the last five years, we have developed the following projects centering on the structural biology of nuclear receptors.<br />
Peroxisome proliferator–activated receptors<br />
The peroxisome proliferator–activated receptors (PPARa, d, and g) are key regulators of glucose and fatty acid homeostasis<br />
and as such are important therapeutic targets for treating cardiovascular disease, diabetes, and cancer. Millions of patients<br />
have benefited from treatment with the PPARg ligands rosiglitazone and pioglitazone for type II diabetes. To understand the<br />
molecular basis of ligand-mediated signaling by PPARs, we have determined crystal structures of each PPAR’s ligand-binding<br />
domain (LBD) bound to many diverse ligands, including fatty acids, the lipid-lowering fibrates, and a new generation of antidiabetic<br />
drugs, the glitazones. We have also determined the crystal structures of these receptors bound to coactivators or<br />
co-repressors, and that of PPARg bound to natural ligand-nitrated fatty acid. These structures provide a framework for understanding<br />
the mechanisms of PPAR agonists and antagonists, as well as the recruitment of coactivators and co-repressors.<br />
We have discovered a number of natural ligands of PPARg. The specific plan of this project is to test the physiological roles<br />
of these PPAR ligands in glucose and insulin regulation, to unravel their molecular and structural mechanisms of action, and to<br />
develop them as therapeutics for diabetes and dislipidemia treatment.<br />
Human glucocorticoid and mineralocorticoid receptors<br />
The human glucocorticoid receptor (GR) and mineralocorticoid receptor (MR) are classic steroid hormone receptors that have<br />
crucial effects on immune/inflammatory responses, metabolic homeostasis, and control of blood pressure. Both GR and MR<br />
are well-established drug targets, and drugs targeting these receptors are sold for more than $10 billion annually. GR ligands<br />
such as dexamethasone (Dex) and fluticasone propionate (FP) are used to treat asthma, leukemia, and autoimmune diseases;<br />
MR ligands such as spironolactone and eplerenone are used to treat hypertension and heart failure. However, the clinical<br />
use of these ligands is limited by undesirable side effects partly associated with their receptor cross-reactivity or low potency.<br />
Thus, the discovery of highly potent and more-selective ligands for GR (such ligands are called “dissociated glucocorticoids”,<br />
which can separate good effects from bad ones) remains an intensive goal of pharmaceutical research.<br />
Recently we determined the structure of GR bound to deacylcortivazol (DAC), which binds to GR with 200-fold more potency<br />
than cortisol, the physiological glucocorticoid. The GR DAC structure reveals that the GR ligand binding pocket can be<br />
expanded dramatically, to twice its normal size. This new pocket provides a tremendous opportunity for drug design and<br />
screening. Using a computational screen, we have identified several nonsteroidal ligands that like dissociated glucocorticoids<br />
in our cell-based assay. We are now running animal studies to confirm the physiological activities of these novel nonsteroidal<br />
ligands, which could lead to new methods of treating inflammation and autoimmune diseases. In addition, we plan to study<br />
the molecular and structural mechanisms of the dissociated glucocorticoids identified by our research.<br />
73
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
The human androgen receptor<br />
The androgen receptor (AR) is the central molecule in the development and progression of prostate cancer. Mutations of<br />
the AR can alter the three-dimensional structure of the receptor in cancer cells and allow the cells to escape the repression<br />
of anti-androgen treatment. In this project, we intend to determine the structural basis of the mutant AR proteins. We<br />
have discovered that AR mutation in prostate cancers often results in enhanced binding to a particular coactivator, SRC-3<br />
(which is also called “amplified in breast cancer 1”, or AIB1). We plan to study the structural and molecular mechanism<br />
of AR antagonists used in prostate cancer treatment and to determine a crystal structure of full-length AR bound to DNA<br />
and coactivator motifs.<br />
Structural genomics of nuclear receptor ligand-binding domains<br />
The LBDs of nuclear receptors contain key structural elements that mediate ligand-dependent regulation of nuclear receptors;<br />
as such, they have been the focus of intense structural study. In the past two years, we have focused on structural characterization<br />
of two orphan receptors: constitutive androstane receptor (CAR) and steroidogenic factor-1 (SF-1), and we<br />
have made significant progress in understanding their ligand binding relationships. In addition, we have identified retinoic<br />
acid as a low-affinity ligand for COUP-TF, which is one of the most conserved nuclear receptors and has essential roles in<br />
angiogenesis, heart development, CNS activity, and metabolic homeostasis. We plan to solve the structures of the remaining<br />
orphan receptors, of which there are only four left.<br />
The Met tyrosine kinase receptor<br />
The MET receptor is a tyrosine kinase that is activated by hepatocyte growth factor/scatter factor (HGF/SF). Aberrant activation<br />
of the Met receptor has been linked with development and metastasis of many types of solid tumors and has been correlated<br />
with poor clinical prognosis. In collaboration with George Vande Woude and Ermanno Gherardi, we plan to develop HGF-Met<br />
antagonists for treating solid tumors.<br />
G protein–coupled receptors<br />
GPCRs form the largest family of receptors in the human genome; they receive signals from photons, ions, small chemicals,<br />
peptides, and protein hormones. Although these receptors account for over 40% of drug targets, their structure remains a<br />
challenge because they are seven-transmembrane receptors. There are only a few crystal structures for class A GPCRs, and<br />
many important questions regarding GPCR ligand binding and activation remain unanswered. Currently my group is focused<br />
on Class B GPCRs, which includes receptors for parathyroid hormone (PTH), corticotropin-releasing factor (CRF), glucagon,<br />
and glucagon-like peptide 1. We have determined crystal structures of the ligand binding domain of the PTH and CRF<br />
receptors, and we are developing hormone analogs for treating osteoporosis, depression, and diabetes. In addition, we are<br />
developing a mammalian overexpression system and plan to use this system for expressing full-length GPCRs for crystallization<br />
and structure studies.<br />
From left, standing:<br />
Zhi, Jurecky, Holtrop, Pioszak, Guthrey, Powell,<br />
Kovach, Melcher, Parker, Bandyopadhyay,<br />
Zhang, E. Xu, Tolbert, Li, Ke, Ruivo<br />
kneeling:<br />
Malapaka, Y. Xu, Reynolds, Zhou, He<br />
74
VARI | <strong>2009</strong><br />
External Collaborators<br />
Eugene Chen and Doug Engel, University of Michigan, Ann Arbor<br />
Bruce Freeman, University of Pittsburgh School of Medicine, Pennsylvania<br />
Thomas J. Gardella, Massachusetts General Hospital and Harvard Medical School<br />
Ermanno Gherardi, University of Cambridge, London, United Kingdom<br />
Steve Kliewer and David Mangelsdorf, University of Texas Southwestern Medical Center, Dallas<br />
Dan R. Littman, New York University School of Medicine, New York<br />
Donald MacDonnell, Duke University, Durham, North Carolina<br />
Clay F. Semenkovich, Washington University, St. Louis, Missouri<br />
Stoney Simmons, National Institutes of Health, Bethesda, Maryland<br />
Scott Thacher, Orphagen Pharmaceuticals, San Diego, California<br />
Brad Thompson and Raj Kumar, University of Texas Medical Branch at Galveston<br />
Ming-Jer Tsai and Sophia Tsai, Baylor College of Medicine, Houston, Texas<br />
Jiemin Wong, Eastern China Normal University, Shanghai<br />
Eu Leong Yong, National University of Singapore<br />
Pfizer Pharmaceuticals<br />
Schering-Plough Pharmaceuticals<br />
Recent Publications<br />
Pioszak, Augen A., Naomi R. Parker, Thomas J. Gardella, and H. Eric Xu. <strong>2009</strong>. Structural basis for parathyriod<br />
hormone–related protein binding to the parathyriod hormone receptor and design of conformation-selective peptides.<br />
Journal of Biological Chemistry 284(41): 28382–28391.<br />
Chakravarthy, Manu V., Irfan J. Lodhi, Li Yin, Raghu V. Malapaka, H. Eric Xu, John Turk, and Clay F. Semenkovich.<br />
<strong>2009</strong>. Identification of a physiologically relevant endogenous ligand for PPARa in liver. Cell 138(3): 467–488.<br />
Knudsen, Beatrice S., Ping Zhao, James Resau, Sandra Cottingham, Ermanno Gherardi, Eric Xu, Bree Berghuis,<br />
Jennifer Daugherty, Tessa Grabinski, Jose Toro, et al. <strong>2009</strong>. A novel multipurpose monoclonal antibody for evaluating<br />
human c-Met expression in preclinical and clinical settings. Applied Immunohistochemistry and Molecular Morphology<br />
17(1): 56–67.<br />
Wang, Zhu, X. Edward Zhou, Daniel L. Motola, Xin Gao, Kelly Suino-Powell, Aoife Conneely, Craig Ogata, Kamalesh K. Sharma,<br />
Richard J. Auchus, James B. Lok, et al. <strong>2009</strong>. Identification of the nuclear receptor DAF-12 as a therapeutic target in<br />
parasitic nematodes. Proceedings of the National Academy of Sciences U.S.A.106(23): 9138–9143.<br />
Kruse, Schoen W., Kelly Suino-Powell, X. Edward Zhou, Jennifer E. Kretschman, Ross Reynolds, Clemens Vonrhein,<br />
Yong Xu, Liliang Wang, Sophia Y. Tsai, Ming-Jer Tsai, and H. Eric Xu. 2008. Identification of COUP-TFII orphan nuclear<br />
receptor as a retinoic acid–activated receptor. PLoS Biology 6(9): e227.<br />
Li, Yong, Jifeng Zhang, Francisco J. Schopfer, Dariusz Martynowski, Minerva T. Garcia-Barrio, Amanda Kovach, Kelly<br />
Suino-Powell, Paul R.S. Baker, Bruce A. Freeman, Y. Eugene Chen, and H. Eric Xu. 2008. Molecular recognition of<br />
nitrated fatty acids by PPARg. Nature Structural & Molecular Biology 15(8): 865–867.<br />
75
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Daniel Nathans Memorial Award<br />
76
VARI | <strong>2009</strong><br />
Daniel Nathans Memorial Award<br />
The Daniel Nathans Memorial Award was established in memory of Dr. Daniel Nathans, a distinguished member of our scientific<br />
community and a founding member of VARI’s Board of <strong>Scientific</strong> Advisors. We established this award to recognize individuals<br />
who emulate Dan and his contributions to biomedical and cancer research. It is our way of thanking and honoring him for his<br />
help and guidance in bringing Jay and Betty Van Andel’s dream to reality. The Daniel Nathans Memorial Award was announced<br />
at our inaugural symposium, “Cancer & Molecular Genetics in the Twenty-First Century”, in September 2000.<br />
Award Recipients<br />
2000 Richard D. Klausner, M.D.<br />
2001 Francis S. Collins, M.D., Ph.D.<br />
2002 Lawrence H. Einhorn, M.D.<br />
2003 Robert A. Weinberg, Ph.D.<br />
2004 Brian Druker, M.D.<br />
2005 Tony Hunter, Ph.D., and Tony Pawson, Ph.D.<br />
2006 Harald zur Hausen, M.D., and Douglas R. Lowy, M.D.<br />
2007 Dennis J. Slamon, M.D., Ph.D., and Genentech, Inc.<br />
From left: Nathans awardees Arthur D. Levinson, Ph.D., representing Genentech, Inc., and Dennis J.<br />
Slamon, M.D., Ph.D., with VARI Director George F. Vande Woude at the Nathans Award ceremony.<br />
77
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Postdoctoral Fellowship Program<br />
78
VARI | <strong>2009</strong><br />
Postdoctoral Fellowship Program<br />
The Van Andel Research Institute provides postdoctoral training opportunities to Ph.D. scientists beginning their research careers.<br />
The fellowships help promising scientists advance their knowledge and research experience while at the same time supporting<br />
the research endeavors of VARI. The fellowships are funded in three ways: 1) by the laboratories to which the fellow is assigned;<br />
2) by the VARI Office of the Director; or 3) by outside agencies. Each fellow is assigned to a scientific investigator who oversees<br />
the progress and direction of research. Fellows who worked in VARI laboratories in 2008 and early <strong>2009</strong> are listed below.<br />
Abhishek Bandyopadhyay<br />
University of Cambridge, United Kingdom<br />
VARI mentor: Eric Xu<br />
Jennifer Bromberg-White<br />
Pennsylvania State University College of<br />
Medicine, Hershey<br />
VARI mentor: Nicholas Duesbery<br />
John Buchweitz<br />
Michigan State University, East Lansing<br />
VARI mentor: Brian Haab<br />
Kathryn Eisenmann<br />
University of Minnesota, Minneapolis<br />
VARI mentor: Arthur Alberts<br />
Leslie Farber<br />
George Washington University,<br />
Washington, D.C.<br />
VARI mentor: Bin Teh<br />
Quliang Gu<br />
Sun Yat-sen University of Medicine, China<br />
VARI mentor: Brian Cao<br />
Dan Huang<br />
Peking Union Medical College, China<br />
VARI mentor: Bin Teh<br />
Schoen Kruse<br />
University of Colorado, Boulder<br />
VARI mentor: Eric Xu<br />
Leanne Lash-Van Wyhe<br />
University of Texas Medical Branch,<br />
Galveston<br />
VARI mentor: Arthur Alberts<br />
Yan Li<br />
Peking Union Medical College, China<br />
VARI mentor: Bin Teh<br />
Brendan Looyenga<br />
University of Michigan, Ann Arbor<br />
VARI mentor: James Resau<br />
Xu Lu<br />
University of Texas Health Sciences Center,<br />
San Antonio<br />
VARI mentor: Steven Triezenberg<br />
Venkata Malapaka<br />
Western Michigan University, Kalamazoo<br />
VARI mentor: Eric Xu<br />
Daisuke Matsuda<br />
Kitasato University, Japan<br />
VARI mentor: Bin Teh<br />
Aikseng Ooi<br />
University of Malaya, Kuala Lumpur<br />
VARI mentor: Bin Teh<br />
Electa Park<br />
Louisiana State University Health Sciences<br />
Center, New Orleans<br />
VARI mentor: Cindy Miranti<br />
Augen Pioszak<br />
University of Michigan, Ann Arbor<br />
VARI mentor: Eric Xu<br />
Dorine Savreux<br />
Virology University, France<br />
VARI mentor: Michael Weinreich<br />
Yi-Mi Wu<br />
National Tsin-Hua University, Taiwan<br />
VARI mentor: Brian Haab<br />
Yong Xu<br />
Shanghai Institute of Materia Medica,<br />
China<br />
VARI mentor: Eric Xu<br />
Chenghai Zhang<br />
Virus Institute of the CDC, China<br />
VARI mentor: Eric Xu<br />
Alex Zhong<br />
Sun Yat-sen University, Guangzhou, China<br />
VARI mentor: Bart Williams<br />
Xiaoyin Zhou<br />
University of Alabama – Birmingham<br />
VARI mentor: Eric Xu<br />
From left: Wu, Park, Lash-Van Wyhe, Ooi, Bandyopadhyay, Bromberg-White, Malapaka, Lu, Pioszak, Zhang, Looyenga, Huang, Zhou, Xu<br />
79
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Student Programs<br />
80
VARI | <strong>2009</strong><br />
Grand Rapids Area Pre-College Engineering Program<br />
The Grand Rapids Area Pre-College Engineering Program (GRAPCEP) is administered by Davenport University and jointly<br />
sponsored and funded by Schering Plough and VARI. The program is designed to provide selected high school students,<br />
who have plans to major in science or genetic engineering in college, with the opportunity to work in a research laboratory. In<br />
addition to research methods, the students also learn workplace success skills such as teamwork and leadership. The four<br />
2008 GRAPCEP students were<br />
Chris Fletcher (Hay/Vande Woude)<br />
Creston High School<br />
Rebecca O’Leary (Resau/Duesbery)<br />
Creston High School<br />
Elisa Van Dyke (Hay/Vande Woude)<br />
Creston High school<br />
Allison Vander Ploeg (Resau/Duesbery)<br />
Creston High School<br />
From left: Fletcher, Van Dyke, Vander Ploeg, O’Leary<br />
81
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Summer Student Internship Program<br />
The VARI student internships were established to provide college students with an opportunity to work with professional researchers<br />
in their fields of interest, to use state-of-the-art equipment and technologies, and to learn valuable people and presentation<br />
skills. At the completion of the 10-week program, the students summarize their projects in an oral presentation or poster.<br />
From January 2008 to March <strong>2009</strong>, VARI hosted more than 65 students from 24 colleges and universities in formal summer<br />
internships under the Frederik and Lena Meijer Student Internship Program and in other student positions during the year. An<br />
asterisk (*) indicates a Meijer student intern.<br />
Aquinas College, Grand Rapids, Michigan<br />
Kevin Coalter (Resau)<br />
Sara Kunz (Hay/Vande Woude)<br />
Kathleen Pollock* (Hay/Vande Woude)<br />
Audrey Sanders (Williams)<br />
Randi VanOcker (Haab)<br />
Butler University, Indianapolis, Indiana<br />
Kevin Maupin (Haab)<br />
Calvin College, Grand Rapids, Michigan<br />
Cheri Ackerman* (MacKeigan)<br />
Lee Heeringa (Haab)<br />
John Snider (Teh)<br />
Katie Van Drunen (Resau)<br />
DePaul University, Chicago, Illinois<br />
Cassie Schumacher* (Williams)<br />
DePauw University, Greencastle, Indiana<br />
Victoria Hledin (Cao)<br />
Ferris State University, Big Rapids, Michigan<br />
Carrie Fiebig (Haab)<br />
Grand Rapids Community College, Michigan<br />
Sara Ramirez (Resau)<br />
Albert Rodriguez (Alberts)<br />
Grand Valley State University, Allendale, Michigan<br />
Ala’a Abughoush (Hay/Vande Woude)<br />
Erica Bechtel (Miranti)<br />
Janell Carruthers (Resau)<br />
Molly Dobb (Webb)<br />
Eric Graf (Miranti)<br />
Craig Johnson (Furge)<br />
Caitlin May* (Weinreich)<br />
Gary Rajah, Jr. (Miranti)<br />
Jonathan Rawson* (Alberts)<br />
Patrick Richardson (Webb)<br />
Doug Roossien, Jr.* (Teh)<br />
Hope College, Holland, Michigan<br />
Nicole Beuschel (Webb)<br />
Indiana University, Bloomington<br />
Sarah Barney (Resau)<br />
Marquette University, Milwaukee, Wisconsin<br />
Michael Avallone (Teh)<br />
Massachusetts Institute of Technology, Cambridge<br />
Shannon Moran (Duesbery)<br />
Michigan State University, East Lansing<br />
Tim Caldwell (Triezenberg)<br />
Ying-Chou Chen, M.S. (Weinreich)<br />
Michelle Dawes* (Duesbery)<br />
Aaron DeWard, B.S. (Alberts)<br />
Pete Haak, B.S. (Resau)<br />
Sebla Kutluay, B.S. (Triezenberg)<br />
Laura Lamb, B.S. (Miranti)<br />
Chih-Shia Lee, M.S. (Duesbery)<br />
Justyne Matheny* (Triezenberg)<br />
Charles Miller, B.S. (Weinreich)<br />
Michael Shaheen (MacKeigan)<br />
Katie Sian, B.S. (MacKeigan)<br />
Susan Spotts, B.S. (Miranti)<br />
Rachel Talaski* (Xu)<br />
Jelani Zarif, M.S. (Miranti)<br />
Nanjing Medical University, China<br />
Guipeng Ding (Cao)<br />
Northern Illinois University, DeKalb<br />
Katsuo Hisano (Resau)<br />
Northern Michigan University, Marquette<br />
Jessica Karasiewicz* (Cao)<br />
Sun Yat-sen University, Guangzhou, China<br />
Rui Sun (Cao)<br />
University of Bath, United Kingdom<br />
Cee Wah Chen (Xu)<br />
Aoife Conneely (Xu)<br />
Fraser Holleywood (Miranti)<br />
University of Dayton, Ohio<br />
Jim Fitzgerald (Teh)<br />
University of Mannheim, Germany<br />
Katja Strunk (Alberts)<br />
University of Michigan, Ann Arbor<br />
Stephanie Berry (Williams)<br />
Xiang Gao (Xu)<br />
Theresa Gipson* (Furge)<br />
Dan Hekman* (Haab)<br />
Hailey Hines (Webb)<br />
Jimmy Hogan (MacKeigan)<br />
Dan Overbeek* (Cavey)<br />
Kyle VanKoevering (Williams)<br />
University of Notre Dame, South Bend, Indiana<br />
Kristin Buzzitta (Teh)<br />
82
VARI | <strong>2009</strong><br />
Summer interns 2008: 1. Cheri Ackerman; 2. Dani Burgenske; 3. Dan Hekman; 4. Jim Fitzgerald; 5. John Snider; 6. Tim Caldwell;<br />
7. Chris Fletcher; 8. Jessica Karasiewicz; 9. Xiang Gao; 10. Randi VanOcker; 11. Jimmy Hogan; 12. Caitlin May;<br />
13. Justyne Matheny; 14. Stephanie Berry; 15. Cassie Schumacher; 16. Theresa Gipson; 17. Nicole Beuschel;<br />
18. Bess Conners; 19. Rebecca O’Leary; 20. Rachel Talaski; 21. Elisa Van Dyke; 22. Josh Van Alstyne;<br />
23. Victoria Hleden; 24. Katie Van Drunen; 25. Leanne Day; 26. Doug Roossien, Jr.; 27. Mitchell Zoerhoff;<br />
28. Chris Cleasby; 29. Allison Vander Ploeg; 30. Dan Overbeek; 31. Michael Shaheen; 32. Naveen Reddy;<br />
33. Kathleen Pollock; 34. Brett Butler; 35. Jonathan Rawson.<br />
University of Wisconsin – Madison<br />
Dani Burgenske* (Resau)<br />
Wellesley College, Wellesley, Massachusetts<br />
Bess Connors (Webb)<br />
Other Van Andel Institute interns<br />
Davenport University, Grand Rapids, Michigan<br />
Sara Hop (Development)<br />
Josh Van Alstyne (Information Technology)<br />
Ferris State University, Big Rapids, Michigan<br />
Chris Cleasby (Facilities)<br />
Leanne Day (General Counsel)<br />
Grand Valley State University, Allendale, MIchigan<br />
Brett Butler (Information Technology)<br />
Naveen Reddy (Business Development)<br />
University of Michigan, Ann Arbor<br />
Mitchell Zoerhoff (Finance)<br />
83
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Han-Mo Koo Memorial Seminar Series<br />
84
VARI | <strong>2009</strong><br />
Han-Mo Koo Memorial Seminar Series<br />
This seminar series is dedicated to the memory of Dr. Han-Mo Koo, who was a VARI <strong>Scientific</strong> Investigator from 1999 until his<br />
passing in May of 2004.<br />
February 2008<br />
David N. Zacks, University of Michigan<br />
“Photoreceptor survival during disease: life hanging in the balance”<br />
Anthony J. Senagore, Spectrum Health<br />
“Economic issues in translational medicine: laparoscopic colectomy”<br />
March<br />
Robert M. Strieter, University of Virginia<br />
“CXC chemokines in angiogenesis and metastases of cancer”<br />
Graham J. Burton, University of Cambridge<br />
“Trophoblast invasion in human pregnancy: functions, mechanisms, and regulation”<br />
Valeri I. Vasioukhin, Fred Hutchinson Cancer Research Center<br />
“Mechanisms of prostate cancer initiation and progression”<br />
April<br />
Anirban Maitra, Johns Hopkins University School of Medicine<br />
“New therapeutic targets for pancreatic cancer”<br />
Patricia E. Fast, International AIDS Vaccine Initiative<br />
“HIV prevention with vaccines and other new prevention technologies:<br />
where do we stand in 2008?”<br />
John L. Cleveland, Scripps Research Institute<br />
“Checkpoints in Myc-induced lymphomagenesis”<br />
Cory Abate-Shen, Columbia University<br />
“Targeting differentiation pathways in mouse models of prostate and bladder cancer”<br />
85
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
June<br />
Donald J. Tindall, Mayo Clinic<br />
“Androgen regulation of gene expression in prostate cancer”<br />
Terry W. Du Clos, University of New Mexico<br />
“C-reactive protein, receptors, ligands, and autoimmunity”<br />
David B. Solit, Memorial Sloan-Kettering Cancer Center<br />
“Genetic predictors of BRAF/MEK-dependence in human tumors”<br />
Kim Orth, University of Texas Southwestern Medical Center<br />
“Black death, black spot, black pearl: dissecting the targets of pathogenic effectors”<br />
July<br />
Brian I. Rini, Case Western Reserve University<br />
“BEGF-targeted therapy in metastatic RCC”<br />
August<br />
Ormond A. MacDougald, University of Michigan<br />
“Role of Wnt signaling in adipose tissues”<br />
Bill M. Bement, University of Wisconsin–Madison<br />
“A Rho GTPase signal treadmill”<br />
September<br />
Bruce H. Littman, Translational Medicine Associates<br />
“The value of translational medicine”<br />
Peter A. Campochiaro, Johns Hopkins Hospital School of Medicine<br />
“Pathogenesis and treatment of ocular neovascularization and excessive vascular permeability”<br />
Sean J. Morrison, University of Michigan<br />
“Stem cell self-renewal and cancer cell proliferation”<br />
86
VARI | <strong>2009</strong><br />
October<br />
Teresa K. Woodruff, Northwestern University<br />
“Regulation of follicle development in vitro and in vivo”<br />
Benjamin G. Neel, Ontario Cancer Institute<br />
“Shp2 mutations in human disease”<br />
Andreas G. Ladurner, European Molecular Biology Laboratory<br />
“The language of chromatin plasticity: identifying new modules and ligands in the regulation of<br />
nucleosome structure”<br />
November<br />
William B. Mattes, Critical Path Institute<br />
“The predictive safety testing consortium: reinventing translational safety assessment through<br />
interdisciplinary and interorganizational collaboration”<br />
Dan R. Littman, New York University Medical Center and Howard Hughes Medical Institute<br />
“Role of the orphan nuclear receptor RORyt in immune system homeostasis”<br />
December<br />
Grant D. Barish, Salk Institute of Biological Studies<br />
“Nuclear receptor and co-repressor control of inflammation: getting SMRT about atherosclerosis”<br />
Dennis J. Slamon, University of California, Los Angeles<br />
Nathans Award Public Lecture: “The diversity of human breast cancer”<br />
Nathans Award <strong>Scientific</strong> Lecture: “Molecular diversity of human breast cancer: clinical and<br />
therapeutic implications”<br />
Arthur D. Levinson, Genentech<br />
Nathans Award Public Lecture: “Cancer biology and the future of personalized medicine”<br />
Nathans Award <strong>Scientific</strong> Lecture: “Herceptin: lessons and prospects for the development of<br />
individualized cancer therapeutics”<br />
Jiandie Lin, University of Michigan<br />
“Metabolic control through the PGC-1 coactivator networks”<br />
87
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
January <strong>2009</strong><br />
Xu Cao, University of Alabama, Birmingham<br />
“TGFb1 as a coupling factor for bone resorption and formation”<br />
February<br />
Tom Mikkelsen, Henry Ford Hospital<br />
“Advances in brain tumor diagnosis and therapy”<br />
M. Arthur Moseley, Duke University<br />
“Gel-free, label-free LC/MS differential expression proteomics: applications at the bench and<br />
at the bedside”<br />
March<br />
John E. Niederhuber, National Cancer Institute<br />
“Cancer as an organ system: the tumor microenviromnent”<br />
88
VARI | <strong>2009</strong><br />
Van Andel Research Institute Organization<br />
89
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
David L. Van Andel,<br />
Chairman and CEO, Van Andel Institute<br />
VARI Board of Trustees<br />
David L. Van Andel, Chairman and CEO<br />
James Fahner, M.D.<br />
Fritz M. Rottman, Ph.D.<br />
Board of <strong>Scientific</strong> Advisors<br />
The Board of <strong>Scientific</strong> Advisors advises the CEO and the Board of Trustees, providing recommendations and suggestions<br />
regarding the overall goals and scientific direction of VARI. The members are<br />
Michael S. Brown, M.D., Chairman<br />
Richard Axel, M.D.<br />
Joseph L. Goldstein, M.D.<br />
Tony Hunter, Ph.D.<br />
Phillip A. Sharp, Ph.D.<br />
<strong>Scientific</strong> Advisory Board<br />
The <strong>Scientific</strong> Advisory Board advises the VARI Director, providing recommendations and suggestions specific to the ongoing<br />
research, especially in the areas of cancer, genomics, and genetics. It also coordinates and oversees the scientific review<br />
process for the Institute’s research programs. The members are<br />
Alan Bernstein, Ph.D.<br />
Joan Brugge, Ph.D.<br />
Webster Cavenee, Ph.D.<br />
Frank McCormick, Ph.D.<br />
90
VARI | <strong>2009</strong><br />
Office of the Director<br />
Jeffrey M. Trent, Ph.D., F.A.C.M.G.<br />
President and Research Director<br />
Deputy Director<br />
for Special Programs<br />
James H. Resau, Ph.D.<br />
Deputy Director<br />
for Research Operations<br />
Nicholas S. Duesbery, Ph.D.<br />
Director<br />
for Research Administration<br />
Roberta Jones<br />
Administrator<br />
to the Director<br />
Michelle Bassett<br />
Science Editor<br />
David E. Nadziejka<br />
Office of the Director Staff<br />
From left: Guthrey, Novakowski,<br />
Koo, Klotz, Minard,<br />
Resau, Nelson, Lewis,<br />
Noyes, Verlin, Patrick<br />
91
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Van Andel Institute Administrative Organization<br />
The organizational units listed below provide administrative support to both the Van Andel Research Institute and the Van Andel<br />
Education Institute.<br />
Executive<br />
David Van Andel, Chairman and CEO<br />
Steven R. Heacock, Chief Administrative Officer and<br />
General Counsel<br />
R. Jack Frick, Chief Financial Officer<br />
Christy Goss, Executive Assistant<br />
Ann Schoen, Executive Assistant<br />
Laura Lohr<br />
Business Development<br />
Jerry Callahan, Ph.D., M.B.A., Vice President<br />
Brent Mulder, Ph.D., M.B.A.<br />
Andrea DeJonge<br />
Thomas DeKoning<br />
Jennifer McGrail<br />
Communications and Development<br />
Joseph P. Gavan, Vice President<br />
Jaime Brookmeyer<br />
Tim Hawkins<br />
Sarah Hop<br />
Sarah Lamb<br />
Gerilyn May<br />
Sarah Smedes<br />
Facilities<br />
Samuel Pinto, Manager<br />
Jeff Cooling<br />
Jason Dawes<br />
Kristi Gentry<br />
Ken De Young<br />
Shelly King<br />
Tracy Lewis<br />
Lewis Lipsey<br />
Dave Marvin<br />
Girlie Peterson<br />
Karen Pittman<br />
Richard Sal<br />
Jose Santos<br />
Richard Ulrich<br />
Pete VanConant<br />
Jeff Wilbourn<br />
Finance<br />
Timothy Myers, Controller<br />
Stephanie Birgy<br />
Cory Cooper<br />
Sandi Dulmes<br />
Richard Herrick<br />
Keri Jackson<br />
Angela Lawrence<br />
Heather Ly<br />
Susan Raymond<br />
Cindy Turner<br />
Jamie VanPortfleet<br />
Theresa Wood<br />
Glassware and Media Services<br />
Richard M. Disbrow, C.P.M., Manager<br />
Bob Sadowski<br />
Marlene Sal<br />
Grants and Contracts<br />
Carolyn W. Witt, Director<br />
Anita Boven<br />
Nicole Doppel<br />
Sara O’Neal<br />
David Ross<br />
Human Resources<br />
Linda Zarzecki, Director<br />
Margie Hoving<br />
Stephanie Koelewyn<br />
Pamela Murray<br />
Angela Plutschouw<br />
92
VARI | <strong>2009</strong><br />
Information Technology<br />
Bryon Campbell, Ph.D., Chief Information Officer<br />
David Drolett, Manager<br />
Bill Baillod<br />
Tom Barney<br />
Phil Bott<br />
Nathan Bumstead<br />
Brad Covell<br />
Charles Grabinski<br />
Kenneth Hoekman<br />
Kimberlee Jeffries<br />
Jason Kotecki<br />
Thad Roelofs<br />
Russell Vander Mey<br />
Candy Wilkerson<br />
Investments Office<br />
Kathleen Vogelsang, Director<br />
Benjamin Carlson<br />
Ted Heilman<br />
Procurement Services<br />
Richard M. Disbrow, C.P.M., Manager<br />
Heather Frazee<br />
Amy Poplaski<br />
John Waldon<br />
Security<br />
Kevin Denhof, CPP, Chief<br />
Andriana Vincent, Team Leader<br />
Amy Davis<br />
Sean Mooney<br />
Maria Straatsma<br />
Chris Wilson<br />
Contract Support<br />
Sarah Lowen, Librarian<br />
(Grand Valley State University)<br />
Jim Kidder, Safety Manager<br />
(Michigan State University)<br />
Logistic Services<br />
Richard M. Disbrow, C.P.M., Manager<br />
Chris Kutschinski<br />
Shannon Moore<br />
93
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
Van Andel Institute<br />
Van Andel Institute Board of Trustees<br />
David Van Andel, Chairman<br />
Peter C. Cook (emeritus)<br />
Ralph W. Hauenstein (emeritus)<br />
Michael Jandernoa<br />
John C. Kennedy<br />
Board of <strong>Scientific</strong> Advisors<br />
Michael S. Brown, M.D., Chairman<br />
Richard Axel, M.D.<br />
Joseph L. Goldstein, M.D.<br />
Tony Hunter, Ph.D.<br />
Phillip A. Sharp, Ph.D.<br />
Van Andel Research Institute<br />
Board of Trustees<br />
David Van Andel, Chairman<br />
James Fahner, M.D.<br />
Fritz M. Rottman, Ph.D.<br />
Van Andel Research Institute<br />
President and<br />
Research Director<br />
Jeffrey M. Trent, Ph.D., F.A.C.M.G.<br />
Chief Administrative Officer<br />
and General Counsel<br />
Steven R. Heacock<br />
Chief Executive Officer<br />
David Van Andel<br />
VP Communications<br />
and Development<br />
Joseph P. Gavan<br />
Van Andel Education Institute<br />
Board of Trustees<br />
David Van Andel, Chairman<br />
Donald W. Maine<br />
Juan R. Olivarez, Ph.D.<br />
Gordon Van Harn, Ph.D.<br />
Gordon Van Wylen, Sc.D.<br />
Van Andel Education Institute<br />
Director<br />
Steven J. Triezenberg, Ph.D.<br />
Chief Financial Officer<br />
R. Jack Frick<br />
94
VARI | <strong>2009</strong><br />
Van Andel Research Institute<br />
PRESIDENT AND RESEARCH DIRECTOR – Jeffrey M. Trent, Ph.D., F.A.C.M.G.<br />
Deputy Directors<br />
Special Programs James Resau, Ph.D.<br />
Research Operations Nick Duesbery, Ph.D.<br />
Director for Research Administration<br />
Roberta Jones<br />
SCIENTIFIC ADVISORY BOARD<br />
Alan Bernstein, Ph.D.<br />
Joan Brugge, Ph.D.<br />
Webster Cavenee, Ph.D.<br />
Frank McCormick, Ph.D.<br />
BASIC SCIENCE<br />
SPECIAL PROGRAMS<br />
Cancer Cell Biology<br />
Brian Haab, Ph.D.<br />
Cancer Immunodiagnostics<br />
George Vande Woude, Ph.D.<br />
Molecular Oncology<br />
Craig Webb, Ph.D.<br />
Tumor Metastasis & Angiogenesis<br />
Signal Transduction<br />
Art Alberts, Ph.D.<br />
Cell Structure & Signal Intergration<br />
Cindy Miranti, Ph.D.<br />
Integrin Signaling & Tumorigenesis<br />
DNA Replication & Repair<br />
Michael Weinreich, Ph.D.<br />
Chromosome Replication<br />
Animal Models<br />
Nicholas Duesbery, Ph.D.<br />
Cancer & Developmental Cell Biology<br />
Bart Williams, Ph.D.<br />
Cell Signaling & Carcinogenesis<br />
Cancer Genetics<br />
Bin Teh, M.D., Ph.D.<br />
Cancer Genetics<br />
Structural Biology<br />
Eric Xu, Ph.D.<br />
Structural Sciences<br />
Systems Biology<br />
Jeffrey MacKeigan, Ph.D.<br />
Systems Biology<br />
Gene Regulation<br />
Steven Triezenberg, Ph.D.<br />
Transcriptional Regulation<br />
Dean of VAI Graduate School<br />
Brian Cao, M.D.<br />
Antibody Technology<br />
Pamela Swiatek, Ph.D., M.B.A.<br />
Germline Modification<br />
& Cytogenetics<br />
Bryn Eagleson, B.S, RLATG<br />
Transgenics and Vivarium<br />
Bin Teh, M.D., Ph.D.<br />
Sequencing<br />
Art Alberts, Ph.D.<br />
Flow Cytometry<br />
Division of Quantitative Sciences<br />
James Resau, Ph.D.<br />
James Resau, Ph.D.<br />
Analytical, Cellular,<br />
& Molecular MIcroscopy<br />
James Resau, Ph.D.<br />
Microarray Technology<br />
Kyle Furge, Ph.D.<br />
Computational Biology<br />
Greg Cavey, B.S.<br />
Mass Spectrometry &<br />
Proteomics<br />
James Resau, Ph.D.<br />
Molecular Epidemiology<br />
95
Van Andel Research Institute | <strong>Scientific</strong> <strong>Report</strong><br />
The Van Andel Institute and/or its affiliated organizations (VARI and VAEI), through its responsible managers, recruits, hires, upgrades, trains,<br />
and promotes in all job titles without regard to race, color, religion, sex, national origin, age, height, weight, marital status,<br />
disability, pregnancy, or veteran status, except when an accommodation is unavailable or it is a bona fide occupational qualification.<br />
Printed by Wolverine Printing Company<br />
96
333 Bostwick Avenue, N.E., Grand Rapids, Michigan 49503<br />
Phone 616.234.5000 Fax 616.234.5001 www.vai.org