Staff Members of the Institute of Biochemistry, TU - Institut für ...
Staff Members of the Institute of Biochemistry, TU - Institut für ...
Staff Members of the Institute of Biochemistry, TU - Institut für ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Nikkomycin biosyn<strong>the</strong>sis<br />
Nikkomycins are produced by several species <strong>of</strong> Streptomyces and exhibit fungicidal, insecticidal<br />
and acaricidal properties due to <strong>the</strong>ir strong inhibition <strong>of</strong> chitin synthase. Nikkomycins are<br />
promising compounds in <strong>the</strong> cure <strong>of</strong> <strong>the</strong> immunosuppressed, such as AIDS patients, organ<br />
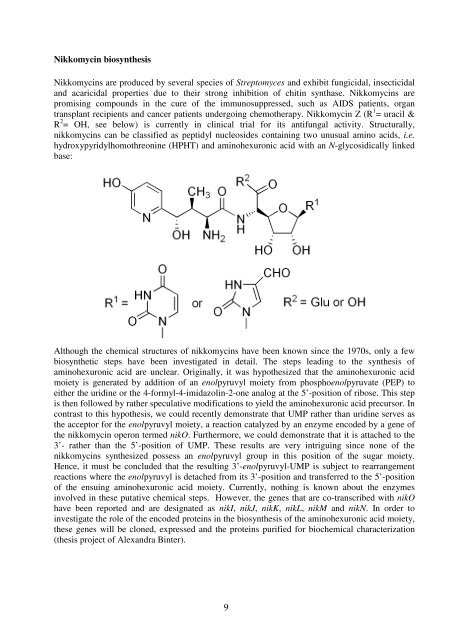
transplant recipients and cancer patients undergoing chemo<strong>the</strong>rapy. Nikkomycin Z (R 1 = uracil &<br />
R 2 = OH, see below) is currently in clinical trial for its antifungal activity. Structurally,<br />
nikkomycins can be classified as peptidyl nucleosides containing two unusual amino acids, i.e.<br />
hydroxypyridylhomothreonine (HPHT) and aminohexuronic acid with an N-glycosidically linked<br />
base:<br />
Although <strong>the</strong> chemical structures <strong>of</strong> nikkomycins have been known since <strong>the</strong> 1970s, only a few<br />
biosyn<strong>the</strong>tic steps have been investigated in detail. The steps leading to <strong>the</strong> syn<strong>the</strong>sis <strong>of</strong><br />
aminohexuronic acid are unclear. Originally, it was hypo<strong>the</strong>sized that <strong>the</strong> aminohexuronic acid<br />
moiety is generated by addition <strong>of</strong> an enolpyruvyl moiety from phosphoenolpyruvate (PEP) to<br />
ei<strong>the</strong>r <strong>the</strong> uridine or <strong>the</strong> 4-formyl-4-imidazolin-2-one analog at <strong>the</strong> 5’-position <strong>of</strong> ribose. This step<br />
is <strong>the</strong>n followed by ra<strong>the</strong>r speculative modifications to yield <strong>the</strong> aminohexuronic acid precursor. In<br />
contrast to this hypo<strong>the</strong>sis, we could recently demonstrate that UMP ra<strong>the</strong>r than uridine serves as<br />
<strong>the</strong> acceptor for <strong>the</strong> enolpyruvyl moiety, a reaction catalyzed by an enzyme encoded by a gene <strong>of</strong><br />
<strong>the</strong> nikkomycin operon termed nikO. Fur<strong>the</strong>rmore, we could demonstrate that it is attached to <strong>the</strong><br />
3’- ra<strong>the</strong>r than <strong>the</strong> 5’-position <strong>of</strong> UMP. These results are very intriguing since none <strong>of</strong> <strong>the</strong><br />
nikkomycins syn<strong>the</strong>sized possess an enolpyruvyl group in this position <strong>of</strong> <strong>the</strong> sugar moiety.<br />
Hence, it must be concluded that <strong>the</strong> resulting 3’-enolpyruvyl-UMP is subject to rearrangement<br />
reactions where <strong>the</strong> enolpyruvyl is detached from its 3’-position and transferred to <strong>the</strong> 5’-position<br />
<strong>of</strong> <strong>the</strong> ensuing aminohexuronic acid moiety. Currently, nothing is known about <strong>the</strong> enzymes<br />
involved in <strong>the</strong>se putative chemical steps. However, <strong>the</strong> genes that are co-transcribed with nikO<br />
have been reported and are designated as nikI, nikJ, nikK, nikL, nikM and nikN. In order to<br />
investigate <strong>the</strong> role <strong>of</strong> <strong>the</strong> encoded proteins in <strong>the</strong> biosyn<strong>the</strong>sis <strong>of</strong> <strong>the</strong> aminohexuronic acid moiety,<br />
<strong>the</strong>se genes will be cloned, expressed and <strong>the</strong> proteins purified for biochemical characterization<br />
(<strong>the</strong>sis project <strong>of</strong> Alexandra Binter).<br />
9