Population dispersion patterns I. Introduction to population dispersion
Population dispersion patterns I. Introduction to population dispersion
Population dispersion patterns I. Introduction to population dispersion
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
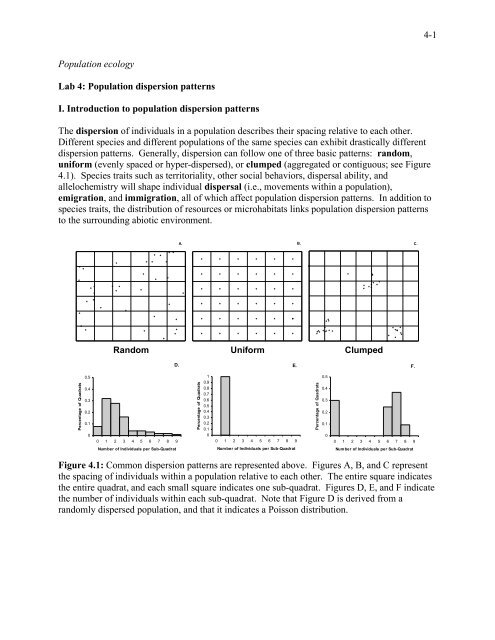
4-1<strong>Population</strong> ecologyLab 4: <strong>Population</strong> <strong>dispersion</strong> <strong>patterns</strong>I. <strong>Introduction</strong> <strong>to</strong> <strong>population</strong> <strong>dispersion</strong> <strong>patterns</strong>The <strong>dispersion</strong> of individuals in a <strong>population</strong> describes their spacing relative <strong>to</strong> each other.Different species and different <strong>population</strong>s of the same species can exhibit drastically different<strong>dispersion</strong> <strong>patterns</strong>. Generally, <strong>dispersion</strong> can follow one of three basic <strong>patterns</strong>: random,uniform (evenly spaced or hyper-dispersed), or clumped (aggregated or contiguous; see Figure4.1). Species traits such as terri<strong>to</strong>riality, other social behaviors, dispersal ability, andallelochemistry will shape individual dispersal (i.e., movements within a <strong>population</strong>),emigration, and immigration, all of which affect <strong>population</strong> <strong>dispersion</strong> <strong>patterns</strong>. In addition <strong>to</strong>species traits, the distribution of resources or microhabitats links <strong>population</strong> <strong>dispersion</strong> <strong>patterns</strong><strong>to</strong> the surrounding abiotic environment.A. B. C.Random Uniform ClumpedD.E.F.Percentage of Quadrats0.50.40.30.20.100 1 2 3 4 5 6 7 8 9Percentage of Quadrats10.90.80.70.60.50.40.30.20.100 1 2 3 4 5 6 7 8 9Percentage of Quadrats0.50.40.30.20.100 1 2 3 4 5 6 7 8 9Number of Individuals per Sub-QuadratNumber of Individuals per Sub-QuadratNumber of Individuals per Sub-QuadratFigure 4.1: Common <strong>dispersion</strong> <strong>patterns</strong> are represented above. Figures A, B, and C representthe spacing of individuals within a <strong>population</strong> relative <strong>to</strong> each other. The entire square indicatesthe entire quadrat, and each small square indicates one sub-quadrat. Figures D, E, and F indicatethe number of individuals within each sub-quadrat. Note that Figure D is derived from arandomly dispersed <strong>population</strong>, and that it indicates a Poisson distribution.
4-2II. Measuring <strong>population</strong> <strong>dispersion</strong><strong>Population</strong> <strong>dispersion</strong> is commonly quantified by <strong>population</strong> ecologists. With mobile organisms,this requires intensive sampling; therefore, we will measure the <strong>dispersion</strong> <strong>patterns</strong> of less mobilespecies. Analyses of <strong>population</strong> <strong>dispersion</strong> <strong>patterns</strong> usually follow a standard method in whichobserved <strong>patterns</strong> are compared <strong>to</strong> predicted, random <strong>dispersion</strong> <strong>patterns</strong> modeled on the Poissondistribution. Deviations from the predicted, random pattern suggest that the <strong>population</strong> understudy exhibits either a uniform or clumped <strong>dispersion</strong> pattern. In <strong>to</strong>day’s lab exercise, we willutilize two different techniques <strong>to</strong> characterize the <strong>dispersion</strong> pattern of our focal species: (1) aquadrat-based method and (2) a point-<strong>to</strong>-plant method.Quadrat MethodThe quadrat method involves counting the frequency of occurrences of the species of interest ineach of the 100 individual 10 X 10 cm sub-quadrats that compose the 1 m 2 quadrat. If theindividuals within the <strong>population</strong> are randomly dispersed, there will be a random number ofindividuals in each quadrat, centered about the mean (see Figure 4.1 A & D). If the individualsin the <strong>population</strong> are uniformly dispersed, there will be the same number of individuals in eachsub-quadrat (see Figure 4.1 B & E). If the individuals in the <strong>population</strong> are clumped in<strong>dispersion</strong>, there will be a few quadrats with many individuals, and many quadrats with noindividuals (see Figure 4.1 C & F).To analyze the data from the quadrat method we will use a chi-square test of hypothesis. Thechi-square test compares a given distribution <strong>to</strong> the Poisson distribution. We will use anequation <strong>to</strong> generate a Poisson distribution with the characteristics that we would expect from arandomly dispersed plant species that has a mean number of plants per sub-quadrat equal <strong>to</strong> oursample <strong>population</strong>. This equation is called the ‘Poisson expression’ by Cox (2001), and it lookslike this:…where e = the base of the natural log = 2.7182818, µ = mean, and x = the frequency category.In Excel, the formula would look like this:=(µ^x)/((EXP(µ))*(FACT(x)))For example, we sample 40 sub-quadrats/cells. Nine cells have 0 individuals, 22 have 1individual, 6 have 2, 2 have 3, 1 has 4 and none of the quadrats/cells have 5 or more individuals(Table 1). Given these values, we can calculate the Poisson probability P(x i ) for each category.
4-3Table 1: Example data -- there are 40 <strong>to</strong>tal sub-quadrats, 44 <strong>to</strong>tal individuals, and a mean of 1.1individuals per sub-quadrat.Number ofIndividuals perSub-Quadrat(x i )Number ofSub-Quadrats(f i )f i x i0 9 9 * 0 = 91 22 1* 22 = 222 6 2 * 6 = 123 2 3 * 2 = 64 1 4 * 1 = 4≥5 0 5 * 0 = 0Σ 40 441.1To calculate the mean value for data in this format use the following equation:…where f is the number of sub-quadrats and x is the number of individuals per sub-quadrat foreach row in Table 1. We can use the Poisson probabilities <strong>to</strong> generate expected probabilitieswith which we can calculate expected values for each row in Table 1.We can then use these expected probabilities <strong>to</strong> calculate expected values using the equationbelow (essentially, multiply each probability above by the <strong>to</strong>tal number of quadrats, in this case,40.):Use these expected values <strong>to</strong> compare with our observed values using a chi-square test. The teststatistic for the chi-square test is χ 2 :In this example, the expected values for quadrats with three or more individuals are combined forthe χ 2 analysis because those quadrats have small sample sizes (2 quadrats with 3 individuals, 1quadrat with 4 individuals, and no quadrats with 5 individuals). If the expected frequency forany category is less than 1, you must add them <strong>to</strong>gether, using summed fi values <strong>to</strong> calculate χ 2(see Table 2). You will also need <strong>to</strong> calculate the degrees of freedom <strong>to</strong> locate the χ 2 statistic ona table:df = k-2
4-4…where k is the number of categories remaining after you perform any necessary adding (2 inour example, Table 2). The χ 2 statistic for our example is 5.841 (Table 2). You can look this upon the χ 2 table, or use the following Excel formula <strong>to</strong> get a precise p-value:=CHIDIST(χ 2 ,df)…where χ 2 is the test statistic you calculated and df are the degrees of freedom.Table 2. Example of a Poisson Table -- Asterisk (*) denotes that the categories for 3, 4, and 5individuals per quadrat were combined in<strong>to</strong> one class in order <strong>to</strong> better meet assumptions of theχ 2 test.Number ofObservedindividualsFrequencyper quadrat(O)(x i )P(x)ExpectedFrequencyE (O-E) 2 /E0 9 0.332871 13.315 1.3981 22 0.366158 14.646 3.6922 6 0.201387 8.055 0.5243 2 0.0738424 1 0.020307 3.945* 0.226*5 0 0.004467Σ 40 1 5.841By plotting the observed and expected values from Table 2, we can see that our data conformclosely <strong>to</strong> the Poisson distribution (Graph 1).Graph 1. Graph of example data.
4-5Point-<strong>to</strong>-plant MethodThe point-<strong>to</strong>-plant distance method utilizes a ratio <strong>to</strong> detect deviation from a random <strong>dispersion</strong>pattern. We use this method <strong>to</strong> sample <strong>dispersion</strong> for organisms that cannot easily be sampledusing a 1 m 2 quadrat (like trees or species that occur much more spaced out). To collect theappropriate data, you will haphazardly select a point of origin (by throwing an object of somesort), and then measure the distance from that point <strong>to</strong> the nearest two individuals of the speciesof interest. Each team will measure 10 haphazardly selected points. These data will be used <strong>to</strong>calculate the sample coefficient of aggregation (A):…where n = the number of sample points, and d = the distance from the selected location andtree 1 or 2. The closest tree should be recorded as d 1 .This coefficient of aggregation will always be between 0 and 1, and the expected value of A for arandomly dispersed <strong>population</strong> is 0.5. The z-equation is used <strong>to</strong> determine if A is significantlydifferent from 0.5:…where n = the number of sample points, 0.2887 = the standard deviation of A values for arandomly dispersed <strong>population</strong>. The calculated z is looked up on the z table <strong>to</strong> find a p-value forthe null hypothesis that A is not significantly different from 0.5. You can use excel <strong>to</strong> look up aprecise p-value from the z-table (z is your calculated z-value):=1-NORMSDIST(z)If A is significantly less than 0.5, the <strong>dispersion</strong> is uniform, and if A is significantly greater than0.5, the <strong>dispersion</strong> is aggregated. Note that this is very similar <strong>to</strong> the equation for a t-test fromthe first lab. The z-equation is simply a special version of the t-equation, except that the degreesof freedom are irrelevant because the number of sample points (n) must always be greater than30, and the <strong>population</strong> variance must be known.III. ObjectiveThe field portion of <strong>to</strong>day’s lab will involve collecting data on the <strong>dispersion</strong> pattern of<strong>population</strong>s of a small, herbaceous plant and a large tree species chosen by your TA. Theobjective of Lab 4 is determine if the <strong>dispersion</strong> <strong>patterns</strong> of the <strong>population</strong>s you investigate arerandom, uniform, or clumped.
4-6IV. InstructionsBefore setting out <strong>to</strong> sample your focal species, complete the following pre-field instructions:1) Generate several testable hypotheses as a class that you can test with <strong>to</strong>day’s exercise.2) Discuss how <strong>to</strong> record the 2 different kinds of <strong>dispersion</strong> data. Set up field datasheets for your sampling procedures.3) Divide in<strong>to</strong> groups and work as teams in the field. Work should be divided up so thatall team members get <strong>to</strong> experience each aspect of the exercise.4) Be sure that you have all the field sampling equipment that you will need.5) All field teams should participate in sampling all habitats. Your TA will pool datafrom all teams <strong>to</strong> generate larger datasets for each <strong>population</strong> that you investigated.Use these complete datasets for your analysis.Field instructions:Sampling for the quadrat method will involve 1 m x 1 m quadrats that are divided in<strong>to</strong> 100 subquadratseach 10 cm x 10 cm. Randomly locate your group’s quadrat within the <strong>population</strong>identified by your TA and determine how many individuals of the species indicated by your TAyou find in your quadrat. Record data for each species by counting the number of individuals ofthe given species in all of your 100 sub-quadrats. Keep track of which grid you are counting(e.g., by using numbers <strong>to</strong> label columns and letters <strong>to</strong> label rows, then identifying each grid witha number-letter designation).To perform the point-<strong>to</strong>-plant distance method, locate the required number of random points inthe <strong>population</strong> of interest. Measure the distance from each random point <strong>to</strong> the two nearest treesof the focal species. Once you are finished, you will have two distances (in meters) for eachrandom point: the distance from the point <strong>to</strong> the nearest tree and the distance from the point <strong>to</strong>the next nearest tree. You will use these data <strong>to</strong> calculate the coefficient of aggregation (A) foryour focal <strong>population</strong> in order carry out the z-test <strong>to</strong> determine the <strong>dispersion</strong> pattern of the<strong>population</strong>.
4-7Literature CitedCox, G. W. 2001. General Ecology Labora<strong>to</strong>ry Manual, 8th edition. McGraw-Hill, New York.Further ReadingCornell, H. V. 1982. The notion of minimum distance or why rare species are clumped.Oecologia 52(2):278-280.Zavala-Hurtado, J. A., P. L. Valverde, M. C. Herrera-Fuentes, A. Diaz-Solis. 2000.Influence of leaf-cutting ants (Atta mexicana) on performance and <strong>dispersion</strong> <strong>patterns</strong> ofperennial desert shrubs in an inter-tropical region of Central Mexico. Journal of AridEnvironments 46(1):93-102.