Introduction to Bioinformatics - Computer Science
Introduction to Bioinformatics - Computer Science
Introduction to Bioinformatics - Computer Science
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
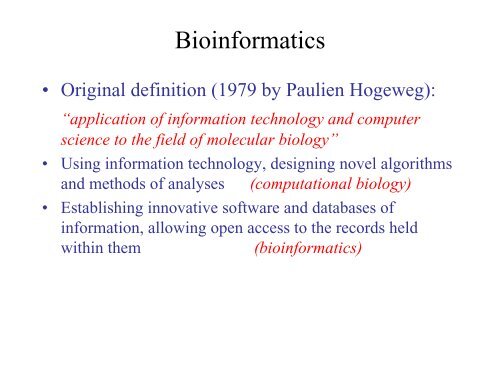
<strong>Bioinformatics</strong> is interdisciplinaryBiomedicineMathematicsStatistics<strong>Computer</strong> <strong>Science</strong>MolecularBiologyStructuralBiology<strong>Bioinformatics</strong>BiophysicsEthical,legal andsocial implicationsEvolutionPatrice Koehl
Central Dogma of Molecular BiologyGenotypeDNAReplicationTranscriptionRNATranslationPhenotypeProteinPatrice Koehl
Genomics: genes give rise <strong>to</strong> proteins• The ~35,000 genes of the human genomeencode > 100,000 polypeptides• Not all of the DNA in a genome encodesproteinmicrobes: 90% coding genehuman: 3% coding gene• About ½ of the non-coding DNA in humans isconserved (functionally important)
DNA structure
RNA structure
Protein structure
Structure Representation (Protein)CPK: hard sphere modelBall-and-stickCar<strong>to</strong>onPatrice Koehl
Central Paradigm of <strong>Bioinformatics</strong>Genomic Sequence Information-> mRNA-> Protein Sequence-> Protein Structure-> Protein Function-> Phenotype
ProteinsSequenceStructureQKPFQCRICMRNFSRSDHLTTHIRTHTG>7million sequences65,000 structures
http://www.ncbi.nlm.nih.gov/genbank/genbankstats.html
Data Mining : Paradigm forScientific Computing• Biology◊ Classifying information anddiscovering unexpectedrelationships◊ EX: Gene ExpressionNetwork◊ Emphasizes: networks,“federated” database
Top ten challenges for bioinformatics1) Precise models of where and when transcriptionwill occur in a genome (initiation and termination)ability <strong>to</strong> predict where and when transcription will occur ingenome2) Precise, predictive models of alternative RNAsplicing: ability <strong>to</strong> predict the splicing pattern of anyprimary transcript in any tissue3) Precise models of signal transduction pathways;ability <strong>to</strong> predict cellular responses <strong>to</strong> external stimuli4) Determining protein:DNA, protein:RNA,protein:protein recognition codes5) Accurate ab-initio protein structure prediction
Top ten challenges for bioinformatics6) Rational design of small molecule inhibi<strong>to</strong>rs ofproteins7) Mechanistic understanding of protein evolution:understanding exactly how new protein functions evolve8) Mechanistic understanding of speciation: moleculardetails of how speciation occurs9) Development of effective gene on<strong>to</strong>logies: systematicways <strong>to</strong> describe gene and protein function10) Education: development of bioinformatics curriculaSource: Birney (EBI), Burge (MIT), Fickett (Glaxo)
Viruses are a subject of study inboth genomics and proteomics• Viruses on the attack:http://www.youtube.com/watch?v=Dh4C-qmfuro&NR=1• Flu Attack! How A Virus Invades Your Body:http://www.youtube.com/watch?v=Rpj0emEGShQ&feature=related• Attacking influenza: how human monoclonalantibodies neutralize the flu virus:http://www.youtube.com/watch?v=lcHy8THENXo&feature=related
A closer look at proteomics:Proteins are central <strong>to</strong> life• DNA codes for proteins• Cellular structure, communications, etc.• Medical, drug development• Failure -> disease (ex: missing, misfolding)• The key <strong>to</strong> protein function is structure
Proteinscollagenimmunoglobulin
The Cycle of LifeStructureSequenceFunctionKKAVINGEQIRSISDLHQTLKKW ELALPEYYGENLDAL WDCLTGVEYPLVLE WRQFEQSKQLTENGAESVLQVFREAKAEGCDITIEvolutionligandFrom Sequence <strong>to</strong> Function and Back…Patrice Koehl
HIERARCHY OF PROTEIN STRUCTURE1.2.3. Tertiary4.
Structure is CentralStructure SpaceProteinStructureDeterminationNMRX-RayNeutron DiffractionPredictionSequence SpaceProteinSequenceDesignDirected evolutionComputational DesignPatrice Koehl
Helices (1)CterNterHydrogen bonds: O (i) N (i+4)Patrice Koehl
Why do proteins fold?Unfolded State-Protein backbone is a linear chain-Chain is self-avoiding-Protein is closely packedFolded State-Amino Acid preferences:- inside (hydrophobic)/ outside (hydrophilic)- Specific interactionsPatrice Koehl
Protein folding and unfoldingNative (0 ns) 1 conformation: compact, order: core, surface.Denatured at higher temperature (8ns, 11ns) flexible, many conformations.Thermal unfolding simulation of 1imf inosi<strong>to</strong>l monophosphataseJohn Hules, National Energy Research Scientific Computing Center (NERSC)http://www.nersc.gov/news/annual_reports/annrep05/research-news/11-proteins.html
Thermodynamic hypothesis: proteinsfall <strong>to</strong> lowest free energy conformationProtein Folding in the Landscape Perspective: Chevron Plots and Non-Arrhenius KineticsHue Sun Chan and Ken A. Dill, Proteins: Structure, Function, and Genetics, 30:1Free energy (vertical axis) as a function of conformation. The twohorizontal axes represent the many chain degrees of freedom.
Energy Landscape1 ms <strong>to</strong> 1sBarrier crossing time~ exp[Barrier Height]1 µsBarrierHeightUnfolded StateExpanded, disorderedMolten GlobuleCompact, disorderedNative StateCompact, OrderedPatrice Koehl
Protein: Linear chain of amino acidsLeuHCαNHHNCOCαOCNHCαSerHNC CαOCOOTrpLys
Degrees of Freedom (DOF) in a protein• Could represent each a<strong>to</strong>m of the side-chain in 3-space.• But a<strong>to</strong>ms, bond lengths and angles close <strong>to</strong> standard foran amino acid: so instead represent in <strong>to</strong>rsion space• TRP (7 side-chain a<strong>to</strong>ms): 21 coords in x,y,z -> 2 coords in X 1 , X 2Patrice Koehl, UC Davis, http://koehllab.genomecenter.ucdavis.edu/teaching
2 different side-chain conformations in LYS2 conformations of lysine side-chain branching of from main chain:only difference is side-chain <strong>to</strong>rsion angles (chi1, chi2, chi3)Left: LYS (-174.0, 70.0, -174.0) Right: LYS (-70.0, -179.0,180.0)
Observe that this <strong>to</strong>rsionmovement removed clashesresulting in lower (more favorable) energy
Optimization problem: find global energyminimum of the protein in <strong>to</strong>rsion spaceEnergy of d1aqr_12: vary res1 (GLN) only105energy100959085100-10595-10090-9585-9080-8575-8070-75807570Depicted here is energy as a function of <strong>to</strong>rsion angles,varying <strong>to</strong>rsion in one side-chain, the rest held constant
Side view of same: observe lots of local minima,even when only one side-chain varies.Energy of d1aqr_12: vary res1 (GLN) only14013012011010090energy130-140120-130110-120100-11090-10080-9070-808070
Designing a Method for PredictingSide-chain Conformation• degrees of freedom• scoring• sampling the search space ofconformations
Search Algorithm -> Sampling• Dead End Elimination• Monte Carlo• Integer Linear Programming• Graph Algorithms• Mean Field Theory
Exhaustive search with <strong>to</strong>y proteinLysLeu3 residues: 9 Leu * 3 Ser * 81 Lys = 2,187 possible conformations.Ser
One way <strong>to</strong> simplify searchLysLeuSerSCMF: Multicopy protein, search in probability space
Protein Docking ProblemInterface: 1AKJ ligand (green) and recep<strong>to</strong>r (blue):