sąrašas - Panevėžio rajono savivaldybė
sąrašas - Panevėžio rajono savivaldybė
sąrašas - Panevėžio rajono savivaldybė
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
EMBL &cetera Issue 11 - July 2002ARP/wARP accelerates at Hamburg thanks to boost from the NIHThe ARP/wARP project at the Hamburg Outstation has justbeen awarded a grant from the NIH which will provide$700,000 over the next four years to further develop the software.ARP/wARP was designed by Victor Lamzin in Hamburg andAnastassis Perrakis, at the National Cancer Institute (NKI) inAmsterdam and formerly of the Grenoble Outstation. Firstreleased in 1998, the software automates the process of buildingmolecular structures from X-ray diffraction patterns. It is beingused in over seven hundred laboratories throughout the world.Creating a new structure depends on relating data from a newmolecule to an extremely accurate, pre-existing model. "For proteincrystals that diffract to two angstroms or lower, which todayis a normal modest resolution, this is a tedious and time-consumingprocess, Victor says. "It requries building a model that is oftensubjective and relies heavily on a great deal of experience on thepart of the user. A few years ago it took Peter Metcalf half a yearto refine a structure. It now takes ARP/wARP half an hour to dothat same job."Improvements in the technology surrounding crystallographyhave opened the field to many more users; researchers with lessexperience are coming to the beamlines with their protein crystals.And structural genomics initiatives hope to dramaticallyincrease the number of structures that will be solved in the nextfew years. The NIH and the European Union are backing theseefforts with significant amounts of funding. EMBL's Outstationsin Hamburg (at DESY) and in Grenoble (at the ESRF), as well asresearch groups at the NKI, recently received grants to participatein an EC-funded project called SPINE. Shortcuts and automatedapproaches such as ARP/wARP will be a necessary componentof high-throughput work at the beamlines."We currently have a relatively redundant collection of about17,000 structures in the protein databases," says CharlesEdmonds, Programme Administrator at the National Institutes ofGeneral Medical Sciences; Edmonds is responsible for a portfolioof grants funded by the NIGMS for technology development tosupport the Protein Structure Initiative (seewww.nigms.nih.gov/funding/psi.html). "Our goal is to collect10,000 more non-redundant structures over the next ten years,with a focus on filling out the catalogue of known protein structurefamilies. Currently we are putting an emphasis on the developmentof technical approaches, and in about three years we planto move more to a production phase."The proposal to continue ARP/wARP's development receivedwhat Edmonds calls "resounding success" in peer review. "It's anexample of investigators who have initiated really originalresearch in areas that we think are unique and important," hesays. "This was obviously critical in securing NIH funding for aproject that doesn't include any American researchers."Lamzin and Perrakis plan to refine and extend the capabilities ofthe software. They plan to improve ARP/wARP's ability to dealwith structural information at lower resolutions but also to makeit easier to use by non-experts. About half of the structures thatare currently determined lie between 2.2 to 2.3 Angstroms in resolution.About two-thirds of existing protein structures have been determinedto a resolution of around 2.3 Å or higher. At this level, theindividual atoms that make up the structure can be distinguishedfairly well from one another. "At that resolution," Lamzin says,"ARP/wARP can deal with the experimental data very well. ThusARP/wARP can be used on about two-thirds of the structuresthat are currently being produced. What we want to achieve is topush the software requirements to the limits – down to a resolutionof 2.7 Angstroms, maybe even 3.0. This will capture at leastnine out of ten of the structures submitted to the protein databank."Come celebrate Hamburg's GroundbreakingCeremony on August 20, 2002!– Russ HodgeThe Hamburg Outstation at DESY is expanding to create a highthroughputcrystallization facility and to give adequate space forour structural genomics activities. All EMBL staff and alumni areinvited to take part in the groundbreaking festivities. Check theHamburg homepage soon for details.– Matthias WilmannsFrench Academy of Sciences honors EMBL Director-General and alumnusEMBL Director General Fotis C. Kafatos has been elected as a foreign associate of the French Academyof Sciences, in the field of "Animal and Plant Biology." In awarding this prestigious honor, theAcademy states: "The work of Fotis Kafatos has relevance for different domains of integrative biology,notably zoology, physiology, ecology, evolution and development, and most recently, parasitology. Hehas played a pioneering role in the development of new concepts and tools in the areas of developmentalbiology, physiology, genetic regulation, and the evolution of complex organisms. He was one of thefirst to introduce molecular biology into the field of development, and made a major discovery in thefield of gene expression during development in a complex organism. In addition, he has played a pioneeringrole in the introduction of genetics, genomics, and cellular biology into the study of the majorvector of malaria. Fotis Kafatos is one of the most respected international biologists due to his contributionsin integrative biology. He has served as an advisor to several French ministers of Research andHigher Education." Fotis received his award at a ceremony at the Academy in JuneIn a separate award, Fotis was also conferred a Docteur Honoris Causa by the Université Louis Pasteur in Strasbourg in March.EMBL Alumnus Riccardo Cortese, who led the Gene Expression Programme during the 1980s, was also named as a Foreign Associateto the French Academy of Sciences this year. The Academy cited his numerous accomplishments, particularly in biomedically-relevantapplied sciences. He left EMBL to found and direct the IRBM (Istituto di Ricerche di Biologia Molecolare) in Pomezia, near Rome.
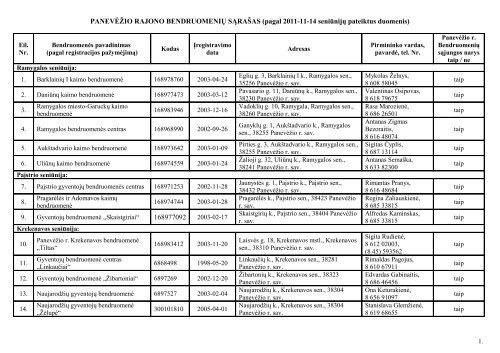
29. Gyventojų bendruomenė „Daukniūnai“ 16897311930. Gyventojų bendruomenė „Piniava“ 168975084 2003-01-3031.32.Gyventojų bendruomenės centras„Šilagalys“Gyventojų bendruomenės centras„Molainiai“168975312 2003-02-05168982125 2003-10-0833. Molainių kaimo bendruomenė 168983184 2003-11-0734. Asociacija „Berniūnų bendruomenė“ 144668 2004-06-0935. Bendruomenės centras „Vaivadai“ 6897656 2003-02-1236.<strong>Panevėžio</strong> <strong>rajono</strong> kaimo bendruomenė„Pažagieniai“6897952 2003-05-1937. Bendruomenė „Savas“ 6897584 2003-02-1038. Kaimo bendruomenė „Senieji Berčiūnai“ 302337859 2009-05-08Naujamiesčio seniūnija:39. Kaimo bendruomenė „Berčiūnai“ 168982844 2003-10-2440. Naujamiesčio piliečių draugija 168940884 1999-06-2841.Gyventojų bendruomenės centras„Dvaras“168968848 2002-09-1742. Kaimo bendruomenė „Gustoniai“ 6897618 2003-02-1143.Gyventojų bendruomenės centras„Liberiškis“Karsakiškio seniūnija:44.<strong>Panevėžio</strong> <strong>rajono</strong> Sodelių kaimobendruomenė45.<strong>Panevėžio</strong> <strong>rajono</strong> Geležių kaimobendruomenė46.<strong>Panevėžio</strong> <strong>rajono</strong> Tiltagalių kaimobendruomenė6897637 2003-02-12168977320 2003-02-246897694 2003-02-17168972355 2002-12-12Sūdramagalos g. 6, Daukniūnų k., <strong>Panevėžio</strong>sen., 38361 <strong>Panevėžio</strong> r. sav.Žaliosios g. 8, Piniavos k., <strong>Panevėžio</strong> sen.,38412 <strong>Panevėžio</strong> r. sav.Kauno g. 2, Šilagalio k., <strong>Panevėžio</strong> sen.,36223 <strong>Panevėžio</strong> r. sav.Karčemos g. 18, Molainių k., <strong>Panevėžio</strong> sen.,37175 <strong>Panevėžio</strong> r. sav.Beržyno g. 24A, Molainių k., <strong>Panevėžio</strong> sen.,37175 <strong>Panevėžio</strong> r. sav.Nevėžio g. 14-5, Berniūnų k., <strong>Panevėžio</strong> sen.,38365 <strong>Panevėžio</strong> r. sav.Žalioji g. 15, Vaivadų k., <strong>Panevėžio</strong> sen.,38447 <strong>Panevėžio</strong> r. sav.Švyturio g. 9-3, Pažagienių k., <strong>Panevėžio</strong>sen., 36222 <strong>Panevėžio</strong> r. sav.Pažagienio g. 40, Pažagienių k., <strong>Panevėžio</strong>sen., 36223 <strong>Panevėžio</strong> r. sav.Berčiūnų k., <strong>Panevėžio</strong> sen., 35290<strong>Panevėžio</strong> r. sav.Rožių g. 1, Berčiūnų k., Naujamiesčio sen.,38357 <strong>Panevėžio</strong> r. sav.Dariaus ir Girėno g. 52, Naujamiesčio mstl.,Naujamiesčio sen., 38335 <strong>Panevėžio</strong> r. sav.Naudvario k., Naujamiesčio sen., 38350<strong>Panevėžio</strong> r. sav.M.Kriaučiūno g. 20, Gustonių k.,Naujamiesčio sen., 38355 <strong>Panevėžio</strong> r. sav.Naujamiesčio g. 1, Liberiškio k.,Naujamiesčio sen., 38330 <strong>Panevėžio</strong> r. sav.Pyvesos g. 50, Sodelių k., Karsakiškio sen.,38458 <strong>Panevėžio</strong> r. sav.Parko g. 2, Geležių mstl., Karsakiškio sen.,38203 <strong>Panevėžio</strong> r. sav.Paežerio g. 2, Tiltagalių k., Karsakiškio sen.,38474 <strong>Panevėžio</strong> r. sav.Reda Matuzevičienė,8 685 43366Zigmantas Babilas,8 698 71240Inga Vapsvienė,8 650 59377Algimantas Lungys,8 698 32145Saulius Gaušas,8 656 74455Sigitas Ulianskas,8 650 39318Valentina Ruzinskienė,8 616 42887Valentinas Geniotis,8 685 47687Nijolė Stankienė,8 606 11924;(8 45) 433914Juozas Mikšys,8 699 87775Marijona Povilonienė,8 614 88424Dalius Dirsė,8 687 19210Irena Andrulienė,8 683 57650Albertas Šilinis,8 699 73635Ona Čeidienė,8 610 12924Vaida Sarapienė,8 698 32562Zita Maršantienė,8 614 86920Laima Baltušnikienė,8 646 07933,8 679 97706taiptaiptaiptaiptaiptaipnetaipnenetaiptaiptaiptaiptaiptaiptaiptaip3.
47.<strong>Panevėžio</strong> <strong>rajono</strong> Karsakiškiobendruomenė48.<strong>Panevėžio</strong> <strong>rajono</strong> Paliūniškio kaimobendruomenėVadoklių seniūnija:49.<strong>Panevėžio</strong> <strong>rajono</strong> Vadoklių ir Mikėnųkaimų bendruomenė50.<strong>Panevėžio</strong> <strong>rajono</strong> Jotainių kaimobendruomenėVelžio seniūnija:51.<strong>Panevėžio</strong> <strong>rajono</strong> kaimo bendruomenė„Maženiai“52.<strong>Panevėžio</strong> <strong>rajono</strong> Kabelių kaimobendruomenė168974025 2003-01-21168977135 2003-01-20168975465 2003-02-07168971449 2002-11-28168984133 2003-12-19168982463 2003-10-2153. Gyventojų bendruomenė „Velžys“ 268978380 2003-04-0854. Dembavos bendruomenė 168976752 2003-02-1455.56.Katinų kaimo gyventojų kaimobendruomenės centrasGyventojų bendruomenės centras„Pastogė“168977288 2003-02-20168970347 2002-10-2957. Liūdynės kaimo bendruomenė 300582500 2006-07-1258. Kaimo bendruomenė „Vyčiai“ 302579086 2010-12-30Lėvens g. 8, Karsakiškio k., Karsakiškio sen.,38462 <strong>Panevėžio</strong> r. sav.Vabalninko g. 15, Paliūniškio k., Karsakiškiosen., 38440 <strong>Panevėžio</strong> r. sav.Statybininkų g. 32, Vadoklių mstl., Vadokliųsen., 38199 <strong>Panevėžio</strong> r. sav.Ramygalos g. 2, Jotainių k., Vadoklių sen.,38218 <strong>Panevėžio</strong> r. sav.Maženių k., Velžio sen., 38134 <strong>Panevėžio</strong> r.sav.Dubų g. 3, Kabelių k., Velžio sen., 38133<strong>Panevėžio</strong> r. sav.Alantos g. 38, Velžio k., Velžio sen., 38128<strong>Panevėžio</strong> r. sav.Veteranų g. 1, Dembavos k., Velžio sen.,38175 <strong>Panevėžio</strong> r. sav.Katinų k., Velžio sen., 38224 <strong>Panevėžio</strong> r.sav.Liūdynės k., Velžio sen., 38130 <strong>Panevėžio</strong> r.sav.Ramioji g. 2, Liūdynės k., Velžio sen., 38130<strong>Panevėžio</strong> r. sav.Zigmo g. 10, Vyčių k., Velžio sen., 38122<strong>Panevėžio</strong> r. sav.Gražina Žemaitienė,8 618 65001Jonas Kvedaravičius,8 682 11894Edita Vanagienė,8 670 93530Milda Kuodienė,8 611 43382Vidmantas Kazlauskas,8 682 97836Algis Petrošaitis,8 687 10957Virginija Stundžienė,8 684 05317Daiva Juodelienė,8 616 52340Genė Vitkevičienė,8 614 90580Romualdas Čepė,8 682 28799Laima Ivanovienė,8 616 98733Kęstutis Brazdžiūnas,8 698 15157taiptaiptaiptaiptaipnetaipnetaiptaiptaipne4.