Core facilities at EMBL â an overview - Molmedrex Project
Core facilities at EMBL â an overview - Molmedrex Project Core facilities at EMBL â an overview - Molmedrex Project
Applications at EMBL• Whole-genome analyses of transcriptome,miRNome & occupancy profiling• Identification of DNA and RNA protein binding sites(ChIP-Seq, CLIP-Seq)• Identification of methylation patterns (Methyl-Seq)• Identification of small ncRNA (miRNA) sequences• Digital gene expression profiling (RNA-Seq)• Whole genome sequencing• De novo sequencing and re-sequencing (pairedendedreads)• Structural variation of genomes – CNV, SNP, in/del
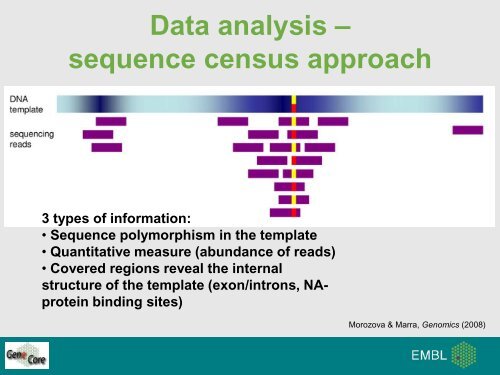
Data analysis –sequence census approach3 types of information:• Sequence polymorphism in the template• Quantitative measure (abundance of reads)• Covered regions reveal the internalstructure of the template (exon/introns, NAproteinbinding sites)Morozova & Marra, Genomics (2008)
- Page 1 and 2: Core facilities at EMBL -an overvie
- Page 3: Research & Core Facilities at EMBLP
- Page 6 and 7: Core Facilities at EMBL• Advanced
- Page 8 and 9: Unlocking the genomeCelniker et al.
- Page 10 and 11: GeneCore ServicesLiquid handling ro
- Page 12 and 13: GeneCore ServicesComplete support &
- Page 14 and 15: Regulation of ERα responsive genes
- Page 16 and 17: EMBL Solexas• ~180 million reads
- Page 18 and 19: Cluster generationPrepare DNAfragme
- Page 22 and 23: Challenges• ChIP-Seq and RNA-Seq
- Page 25 and 26: AcknowledgementRichard CarmoucheJü
D<strong>at</strong>a <strong>an</strong>alysis –sequence census approach3 types of inform<strong>at</strong>ion:• Sequence polymorphism in the templ<strong>at</strong>e• Qu<strong>an</strong>tit<strong>at</strong>ive measure (abund<strong>an</strong>ce of reads)• Covered regions reveal the internalstructure of the templ<strong>at</strong>e (exon/introns, NAproteinbinding sites)Morozova & Marra, Genomics (2008)