Protein Engineering Protocols - Mycobacteriology research center
Protein Engineering Protocols - Mycobacteriology research center Protein Engineering Protocols - Mycobacteriology research center
3Considerations in the Design and Optimizationof Coiled Coil StructuresJody M. Mason, Kristian M. Müller, and Katja M. ArndtSummaryCoiled coil motifs are, despite their apparent simplicity, highly specific, and play a significantrole in the understanding of tertiary structure and its formation. The most commonly observed ofthe coiled coils, the parallel dimeric, is yet to be fully characterized for this structural class in general.Nonetheless, strict rules have emerged for the necessity of specific types of amino acids atspecific positions. In this chapter, we discuss this system in light of existing coiled coil structuresand in applying rules to coiled coils that are to be designed or optimized. Understanding andexpanding on these rules is crucial in using these motifs, which play key roles in virtually everycellular process, to act as drug-delivery agents by sequestering other proteins that are not behavingnatively or that have been upregulated (for example, by binding to coiled coil domains implicatedin oncogenesis). The roles of the a and d “hydrophobic” core positions and the e and g“electrostatic” edge positions in directing oligomerization and pairing specificity are discussed.Also discussed is the role of these positions in concert with the b, c, and f positions in maintainingα-helical propensity, helix solubility, and dimer stability.Key Words: Coiled coil; helix; heptad repeat; in vivo selection; leucine zipper; library design;protein design; protein engineering; protein fragment complementation assay; protein stability;rational design.1. IntroductionThe coiled coil is a common structural motif estimated to constitute 3 to 5%of the encoded residues in most genomes (1). It consists of two to five α-helicesthat habitually twist around each other, typically left-handedly, to form a supercoil.Whereas regular α-helices go through 3.6 residues for each complete turnof the helix, the distortion imposed on each helix within a left-handed coiledcoil lowers this value to 3.5. This equates to a seven amino acid repeat for everytwo turns of the helix (2,3). The most frequently occurring type of coiled coilFrom: Methods in Molecular Biology, vol. 352: Protein Engineering ProtocolsEdited by: K. M. Arndt and K. M. Müller © Humana Press Inc., Totowa, NJ35
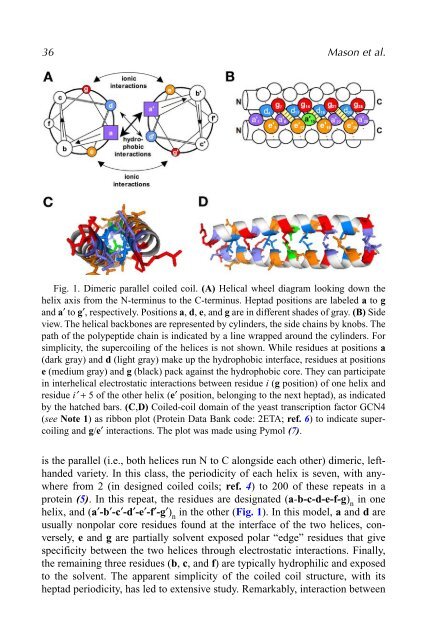
36 Mason et al.Fig. 1. Dimeric parallel coiled coil. (A) Helical wheel diagram looking down thehelix axis from the N-terminus to the C-terminus. Heptad positions are labeled a to gand a′ to g′, respectively. Positions a, d, e, and g are in different shades of gray. (B) Sideview. The helical backbones are represented by cylinders, the side chains by knobs. Thepath of the polypeptide chain is indicated by a line wrapped around the cylinders. Forsimplicity, the supercoiling of the helices is not shown. While residues at positions a(dark gray) and d (light gray) make up the hydrophobic interface, residues at positionse (medium gray) and g (black) pack against the hydrophobic core. They can participatein interhelical electrostatic interactions between residue i (g position) of one helix andresidue i′ + 5 of the other helix (e′ position, belonging to the next heptad), as indicatedby the hatched bars. (C,D) Coiled-coil domain of the yeast transcription factor GCN4(see Note 1) as ribbon plot (Protein Data Bank code: 2ETA; ref. 6) to indicate supercoilingand g/e′ interactions. The plot was made using Pymol (7).is the parallel (i.e., both helices run N to C alongside each other) dimeric, lefthandedvariety. In this class, the periodicity of each helix is seven, with anywherefrom 2 (in designed coiled coils; ref. 4) to 200 of these repeats in aprotein (5). In this repeat, the residues are designated (a-b-c-d-e-f-g) nin onehelix, and (a′-b′-c′-d′-e′-f′-g′) nin the other (Fig. 1). In this model, a and d areusually nonpolar core residues found at the interface of the two helices, conversely,e and g are partially solvent exposed polar “edge” residues that givespecificity between the two helices through electrostatic interactions. Finally,the remaining three residues (b, c, and f) are typically hydrophilic and exposedto the solvent. The apparent simplicity of the coiled coil structure, with itsheptad periodicity, has led to extensive study. Remarkably, interaction between
- Page 1 and 2: METHODS IN MOLECULAR BIOLOGY 352Pr
- Page 4 and 5: M E T H O D S I N M O L E C U L A R
- Page 6 and 7: PrefaceProtein engineering is a fas
- Page 8 and 9: ContentsPreface ...................
- Page 10 and 11: ContributorsKATJA M. ARNDT • Inst
- Page 12: Contributors xiKAZUNARI TAIRA • D
- Page 16 and 17: 1Combinatorial Protein Design Strat
- Page 18 and 19: Combinatorial Protein Design Strate
- Page 20 and 21: Combinatorial Protein Design Strate
- Page 22 and 23: Combinatorial Protein Design Strate
- Page 24 and 25: Combinatorial Protein Design Strate
- Page 26 and 27: Combinatorial Protein Design Strate
- Page 28 and 29: Combinatorial Protein Design Strate
- Page 30 and 31: Combinatorial Protein Design Strate
- Page 32 and 33: Combinatorial Protein Design Strate
- Page 34 and 35: Combinatorial Protein Design Strate
- Page 36 and 37: 2Global Incorporation of Unnatural
- Page 38 and 39: Incorporation of Unnatural Amino Ac
- Page 40 and 41: Incorporation of Unnatural Amino Ac
- Page 42 and 43: Incorporation of Unnatural Amino Ac
- Page 44 and 45: Incorporation of Unnatural Amino Ac
- Page 46 and 47: Incorporation of Unnatural Amino Ac
- Page 50 and 51: Design of Coiled Coil Structures 37
- Page 52 and 53: Design of Coiled Coil Structures 39
- Page 54 and 55: Design of Coiled Coil Structures 41
- Page 56 and 57: Design of Coiled Coil Structures 43
- Page 58 and 59: Design of Coiled Coil Structures 45
- Page 60 and 61: Design of Coiled Coil Structures 47
- Page 62 and 63: Design of Coiled Coil Structures 49
- Page 64 and 65: Design of Coiled Coil Structures 51
- Page 66 and 67: Design of Coiled Coil Structures 53
- Page 68 and 69: Design of Coiled Coil Structures 55
- Page 70 and 71: Design of Coiled Coil Structures 57
- Page 72 and 73: Design of Coiled Coil Structures 59
- Page 74 and 75: Design of Coiled Coil Structures 61
- Page 76 and 77: Design of Coiled Coil Structures 63
- Page 78 and 79: Design of Coiled Coil Structures 65
- Page 80 and 81: Design of Coiled Coil Structures 67
- Page 82 and 83: Design of Coiled Coil Structures 69
- Page 84 and 85: 4Calcium Indicators Based on Calmod
- Page 86 and 87: Protein-Based Ca 2+ Indicators 73Fi
- Page 88 and 89: Protein-Based Ca 2+ Indicators 7512
- Page 90 and 91: Protein-Based Ca 2+ Indicators 77Fi
- Page 92 and 93: Protein-Based Ca 2+ Indicators 79Fi
- Page 94 and 95: Protein-Based Ca 2+ Indicators 8145
- Page 96 and 97: 5Design and Synthesis of Artificial
36 Mason et al.Fig. 1. Dimeric parallel coiled coil. (A) Helical wheel diagram looking down thehelix axis from the N-terminus to the C-terminus. Heptad positions are labeled a to gand a′ to g′, respectively. Positions a, d, e, and g are in different shades of gray. (B) Sideview. The helical backbones are represented by cylinders, the side chains by knobs. Thepath of the polypeptide chain is indicated by a line wrapped around the cylinders. Forsimplicity, the supercoiling of the helices is not shown. While residues at positions a(dark gray) and d (light gray) make up the hydrophobic interface, residues at positionse (medium gray) and g (black) pack against the hydrophobic core. They can participatein interhelical electrostatic interactions between residue i (g position) of one helix andresidue i′ + 5 of the other helix (e′ position, belonging to the next heptad), as indicatedby the hatched bars. (C,D) Coiled-coil domain of the yeast transcription factor GCN4(see Note 1) as ribbon plot (<strong>Protein</strong> Data Bank code: 2ETA; ref. 6) to indicate supercoilingand g/e′ interactions. The plot was made using Pymol (7).is the parallel (i.e., both helices run N to C alongside each other) dimeric, lefthandedvariety. In this class, the periodicity of each helix is seven, with anywherefrom 2 (in designed coiled coils; ref. 4) to 200 of these repeats in aprotein (5). In this repeat, the residues are designated (a-b-c-d-e-f-g) nin onehelix, and (a′-b′-c′-d′-e′-f′-g′) nin the other (Fig. 1). In this model, a and d areusually nonpolar core residues found at the interface of the two helices, conversely,e and g are partially solvent exposed polar “edge” residues that givespecificity between the two helices through electrostatic interactions. Finally,the remaining three residues (b, c, and f) are typically hydrophilic and exposedto the solvent. The apparent simplicity of the coiled coil structure, with itsheptad periodicity, has led to extensive study. Remarkably, interaction between