Indexing Sparse graphs for Similarity Search - IJETTCS ...
Indexing Sparse graphs for Similarity Search - IJETTCS ...
Indexing Sparse graphs for Similarity Search - IJETTCS ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
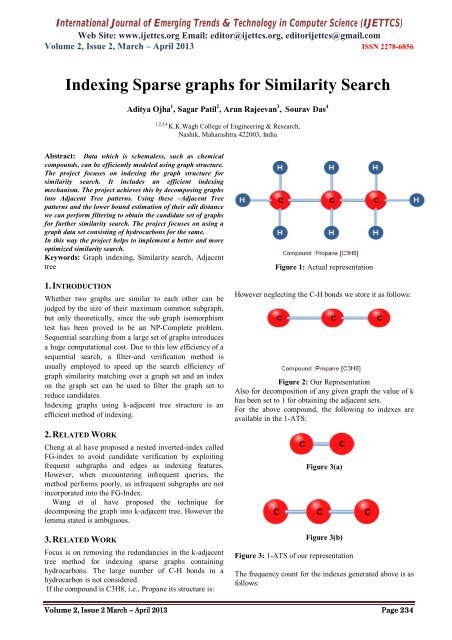
International Journal of Emerging Trends & Technology in Computer Science (<strong>IJETTCS</strong>)Web Site: www.ijettcs.org Email: editor@ijettcs.org, editorijettcs@gmail.comVolume 2, Issue 2, March – April 2013 ISSN 2278-6856<strong>Indexing</strong> <strong>Sparse</strong> <strong>graphs</strong> <strong>for</strong> <strong>Similarity</strong> <strong>Search</strong>Aditya Ojha 1 , Sagar Patil 2 , Arun Rajeevan 3 , Sourav Das 41,2,3,4K.K.Wagh College of Engineering & Research,Nashik, Maharashtra 422003, IndiaAbstract: Data which is schemaless, such as chemicalcompounds, can be efficiently modeled using graph structure.The project focuses on indexing the graph structure <strong>for</strong>similarity search. It includes an efficient indexingmechanism. The project achieves this by decomposing <strong>graphs</strong>into Adjacent Tree patterns. Using these –Adjacent Treepatterns and the lower bound estimation of their edit distancewe can per<strong>for</strong>m filtering to obtain the candidate set of <strong>graphs</strong><strong>for</strong> further similarity search. The project focuses on using agraph data set consisting of hydrocarbons <strong>for</strong> the same.In this way the project helps to implement a better and moreoptimized similarity search.Keywords: Graph indexing, <strong>Similarity</strong> search, Adjacenttree1. INTRODUCTIONWhether two <strong>graphs</strong> are similar to each other can bejudged by the size of their maximum common subgraph,but only theoretically, since the sub graph isomorphismtest has been proved to be an NP-Complete problem.Sequential searching from a large set of <strong>graphs</strong> introducesa huge computational cost. Due to this low efficiency of asequential search, a filter-and verification method isusually employed to speed up the search efficiency ofgraph similarity matching over a graph set and an indexon the graph set can be used to filter the graph set toreduce candidates.<strong>Indexing</strong> <strong>graphs</strong> using k-adjacent tree structure is anefficient method of indexing.2. RELATED WORKCheng at al have proposed a nested inverted-index calledFG-index to avoid candidate verification by exploitingfrequent sub<strong>graphs</strong> and edges as indexing features.However, when encountering infrequent queries, themethod per<strong>for</strong>ms poorly, as infrequent sub<strong>graphs</strong> are notincorporated into the FG-Index.Wang et al have proposed the technique <strong>for</strong>decomposing the graph into k-adjacent tree. However thelemma stated is ambiguous.3. RELATED WORKFocus is on removing the redundancies in the k-adjacenttree method <strong>for</strong> indexing sparse <strong>graphs</strong> containinghydrocarbons. The large number of C-H bonds in ahydrocarbon is not considered.If the compound is C3H8, i.e., Propane its structure is:Figure 1: Actual representationHowever neglecting the C-H bonds we store it as follows:Figure 2: Our RepresentationAlso <strong>for</strong> decomposition of any given graph the value of khas been set to 1 <strong>for</strong> obtaining the adjacent sets.For the above compound, the following to indexes areavailable in the 1-ATS:Figure 3(a)Figure 3(b)Figure 3: 1-ATS of our representationThe frequency count <strong>for</strong> the indexes generated above is asfollows:Volume 2, Issue 2 March – April 2013 Page 234
International Journal of Emerging Trends & Technology in Computer Science (<strong>IJETTCS</strong>)Web Site: www.ijettcs.org Email: editor@ijettcs.org, editorijettcs@gmail.comVolume 2, Issue 2, March – April 2013 ISSN 2278-6856Table 1: Frequency countIndex Structure Frequency CountCC1 2CC1C1 1The <strong>graphs</strong> are evaluated <strong>for</strong> similarity on the basis of thefollowing lemma:Edit Distance:The Graph Edit Distance between two <strong>graphs</strong> G1 and G2is the minimum number of GEOs needed to trans<strong>for</strong>m G1to a graph isomorphic to G2. The definition of editdistance of two <strong>graphs</strong> gives us a measurement toquantify the difference of two <strong>graphs</strong>.Table 3: Frequency Count of 1-ATS in sample graphIndex Structure Frequency CountCC1O2 1CC1C1 1CC1N1 1Here |=2|V(Q)| = 7Hence <strong>for</strong> graph edit distance of 3 both <strong>graphs</strong> would besimilar.The following figure represents a basic block diagram <strong>for</strong>the proposed system.The GEO can be one of the following six operations:1. Delete an edge from the graph.2. Insert an edge between two disconnected vertices.3. Delete an isolated vertex from the graph.4. Insert an isolated vertex into the graph.5. Change the label of a vertex.6. Change the label of an edge.Consider the following two <strong>graphs</strong>:Query Graph:Figure 4: A sample Query GraphTable 2: Frequency count of 1-ATS in Query GraphIndex Structure Frequency CountCC1 1CC1C1 2CC1C1O2 1CC1N1 1Graph in dataset:Figure 5: A sample Graph from the dataset4. CONCLUSIONFigure 6: Block DiagramBy decomposing the <strong>graphs</strong> into small pieces (1-ATs),and pairing-up these pieces, we evaluate the globalsimilarity between them. In order to seek <strong>for</strong> acompromise between frequent-subgraph-based indexingmethods and graph-decomposition-based indexingmethods, we use the redundant subtree structure: 1-ATpattern <strong>for</strong> index construction. 1-AT records morestructural in<strong>for</strong>mation on each vertex than a normalgraph-decomposition- based indexing method, and whilemaintaining the simple structure of tree. By calculatingthe number of common 1-ATs of two <strong>graphs</strong>, we canestimate the graph edit distance between them.This gives us a method <strong>for</strong> indexing and candidatefiltering in a graph set <strong>for</strong> similarity matching.ACKNOWLEDGEMENTWe would like to express our sincere gratitude andappreciation to our project guide Prof. Rutuja Jadhav <strong>for</strong>the patience, guidance, help and <strong>for</strong> being our greatestsource of in<strong>for</strong>mation during this project. We also thankProf. Kamlapur S., <strong>for</strong> providing time and helpfulcomments <strong>for</strong> our work.References[1] T.H. Cormen, “Np Completeness,” Introduction toAlgorithms, W. Yu, ed., second ed., vol. 7, pp. 620-630. China Machine Press, 2007.Volume 2, Issue 2 March – April 2013 Page 235
International Journal of Emerging Trends & Technology in Computer Science (<strong>IJETTCS</strong>)Web Site: www.ijettcs.org Email: editor@ijettcs.org, editorijettcs@gmail.comVolume 2, Issue 2, March – April 2013 ISSN 2278-6856[2] Efficiently <strong>Indexing</strong> Large <strong>Sparse</strong> Graphs <strong>for</strong><strong>Similarity</strong> <strong>Search</strong> Guoren Wang, Bin Wang,Xiaochun Yang, Member, IEEE Computer Society,and Ge Yu, Member, IEEE IEEE TRANSACTIONSON KNOWLEDGE AND DATA ENGINEERING,VOL. 24, NO. 3, MARCH 2012[3] Data Structures and Algorithms in Java (2nd Edition)by Robert La<strong>for</strong>e[4] Introductory Graph Theory by Gary Chartrand[5] Thinking in Java (4th Edition) Bruce Eckel[6] M. Kuramochi and G. Karypis, “Frequent SubgraphDiscovery,” Proc. 2001 IEEE Int’l Conf. DataMining, pp. 313-320, 2001.[7] S. Sarawagi and A. Kirpal, “Efficient Set Joins on<strong>Similarity</strong> Predicates,” Proc. ACM SIGMOD, pp.743-754, 2004.[8] D. Justice and A. Hero, “A Binary LinearProgramming Formulation of the Graph EditDistance,” IEEE Trans. Pattern Analysis andMachine Intelligence, vol. 28, no. 8, pp. 1200-1214,Aug. 2006.[9] O. Johansson, “Graph Decomposition Using NodeLabels,” doctoral dissertation, Royal Inst. ofTechnology, 2001.[10] Y. Tian and J.M. Patel, “Tale: A Tool <strong>for</strong>Approximate Large Graph Matching,” Proc. 24thInt’l Conf. Data Eng., pp. 963-972, 2008.[11] H. Jiang, H. Wang, P.S. Yu, and S. Zhou, “Gstring:A Novel Approach <strong>for</strong> Efficient <strong>Search</strong> in GraphDatabases,” Proc. 23rd Int’l Conf. Data Eng., pp.566-575, 2007.[12] L. Zou, L. Chen, J.X. Yu, and Y. Lu, “A NovelSpectral Coding in a Large Graph Database,” Proc.11th Int’l Conf. Extending DatabaseTechnology, pp.181-192, 2008.[13] D.W.Williams, J. Huan, and W. Wang, “GraphDatabase <strong>Indexing</strong> Using Structured GraphDecomposition,” Proc. 23rd Int’l Conf. Data Eng.,pp. 976-985, 2007.[19] D. Shasha, J.T.-L. Wang, and R. Giugno,“Algorithmics and Applications of Tree and Graph<strong>Search</strong>ing,” Proc. 21st ACMSIGACT-SIGMOD-SIGART Symp. Principles of Database Systems, pp.39-52, 2002.Sourav Das is an U.G. student at KKWIEER,University of Pune. He is a member of CSI. Hisareas of interest are embedded software, P2Pnetworks, data qualityArun Rajeevan is an U.G. student at KKWIEER,University of Pune. His areas of interest are neuralnetworks, databases, digital signal processing.AUTHORAditya Ojha is an U.G. student at KKWIEER,University of Pune. He is also an IBM StudentAmbassador <strong>for</strong> TGMC. He is a member of CSI,his areas of interest are parallel and distributedsystems, database theory, networking.Sagar Patil is an U.G. student at KKWIEER,University of Pune. He is a member of CSI. Hisareas of interest are graph theory, advancedoperating systems, analysis of algorithms.Volume 2, Issue 2 March – April 2013 Page 236