Pawel Bednarek, Michal Jasinski - PolSCA
Pawel Bednarek, Michal Jasinski - PolSCA Pawel Bednarek, Michal Jasinski - PolSCA
Plant RNA virusesBrome Mosaic VirusBrad Bean Mottle VirusRed Clover Necrotic Mosaic VirusTurnip Vein Clearing VirusCucumber Mosaic VirusPeanut Stunt VirusPepino Mosaic VirusTomato Black Ring VirusMethods applied tosearching for therecombinants in viralpopulations.genetic variabilityHasiow-Jaroszewska et.al.(2012) Two types of defective RNAs arising from the tomato black ringvirus genome . Arch.Virol. 157:569–572Sztuba-Solińska et. al.(2011) RNA-RNA recombination in plant virus replication and evolution. Annu. Rev. Phytopathol.49:415-43.Urbanowicz et. al. (2005) Homologous crossovers among molecules of brome mosaic bromovirus RNA1 or RNA2segments in vivo. J. Virol. 79:5732-42.virus-plant interactionsHasiow-Jaroszewska et.al., Single mutation converts mild pathotype of the Pepinomosaic virus into necrotic one. Virus Research (2011) 159: Pages 57–61vectors for protein expressionLocal and systemicexpression of GFPusing transientexpression viralsystem(IconGenetics)
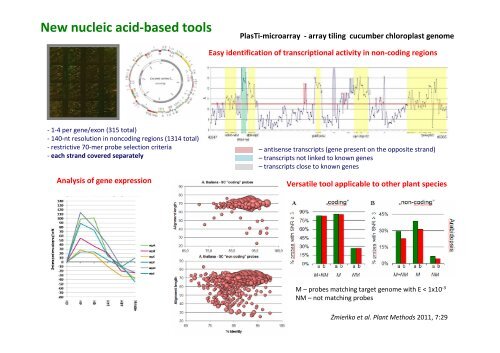
New nucleic acid-based toolsPlasTi-microarray - array tiling cucumber chloroplast genomeEasy identification of transcriptional activity in non-coding regions- 1-4 per gene/exon (315 total)- 140-nt resolution in noncoding regions (1314 total)- restrictive 70-mer probe selection criteria- each strand covered separatelyAnalysis of gene expression– antisense transcripts (gene present on the opposite strand)– transcripts not linked to known genes– transcripts close to known genesVersatile tool applicable to other plant speciesM – probes matching target genome with E < 1x10 -3NM – not matching probesŻmieńko et al. Plant Methods 2011, 7:29
- Page 1 and 2: Institute of Bioorganic ChemistryPo
- Page 3 and 4: IBCH - general information
- Page 5 and 6: IBCHHUMAN RESOURCESOverall employme
- Page 7: 2008Start of European Center of Bio
- Page 11 and 12: Fundamental research:• DNA and RN
- Page 13 and 14: RESEARCH LABORATORIESLaboratory of
- Page 15 and 16: Major research topicsI. RNA in plan
- Page 17: RNA in plant gene regulationStructu
- Page 21 and 22: organism genome size (MB) ABC prote
- Page 23 and 24: • POLAPGEN-BD project financing a
- Page 25 and 26: Trp-derived metabolites are critica
- Page 27: Thank you for your kind attention
New nucleic acid-based toolsPlasTi-microarray - array tiling cucumber chloroplast genomeEasy identification of transcriptional activity in non-coding regions- 1-4 per gene/exon (315 total)- 140-nt resolution in noncoding regions (1314 total)- restrictive 70-mer probe selection criteria- each strand covered separatelyAnalysis of gene expression– antisense transcripts (gene present on the opposite strand)– transcripts not linked to known genes– transcripts close to known genesVersatile tool applicable to other plant speciesM – probes matching target genome with E < 1x10 -3NM – not matching probesŻmieńko et al. Plant Methods 2011, 7:29