Novel genetic and epigenetic alterations in ... - Ous-research.no
Novel genetic and epigenetic alterations in ... - Ous-research.no
Novel genetic and epigenetic alterations in ... - Ous-research.no
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
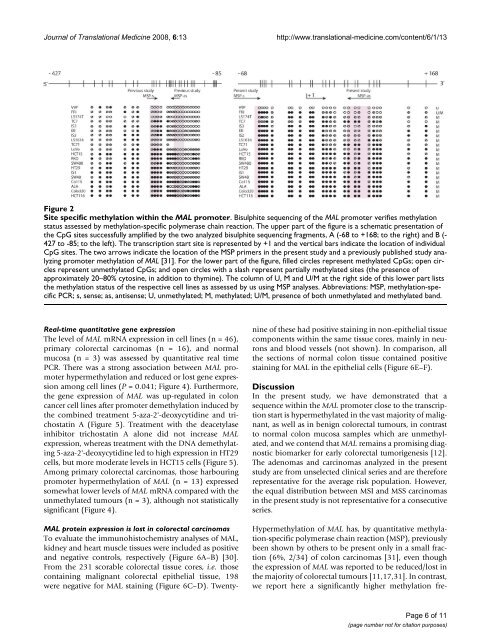
Journal of Translational Medic<strong>in</strong>e 2008, 6:13http://www.translational-medic<strong>in</strong>e.com/content/6/1/13Site specific methylation with<strong>in</strong> the MAL promoterFigure 2Site specific methylation with<strong>in</strong> the MAL promoter. Bisulphite sequenc<strong>in</strong>g of the MAL promoter verifies methylationstatus assessed by methylation-specific polymerase cha<strong>in</strong> reaction. The upper part of the figure is a schematic presentation ofthe CpG sites successfully amplified by the two analyzed bisulphite sequenc<strong>in</strong>g fragments, A (-68 to +168; to the right) <strong>and</strong> B (-427 to -85; to the left). The transcription start site is represented by +1 <strong>and</strong> the vertical bars <strong>in</strong>dicate the location of <strong>in</strong>dividualCpG sites. The two arrows <strong>in</strong>dicate the location of the MSP primers <strong>in</strong> the present study <strong>and</strong> a previously published study analyz<strong>in</strong>gpromoter methylation of MAL [31]. For the lower part of the figure, filled circles represent methylated CpGs; open circlesrepresent unmethylated CpGs; <strong>and</strong> open circles with a slash represent partially methylated sites (the presence ofapproximately 20–80% cytos<strong>in</strong>e, <strong>in</strong> addition to thym<strong>in</strong>e). The column of U, M <strong>and</strong> U/M at the right side of this lower part liststhe methylation status of the respective cell l<strong>in</strong>es as assessed by us us<strong>in</strong>g MSP analyses. Abbreviations: MSP, methylation-specificPCR; s, sense; as, antisense; U, unmethylated; M, methylated; U/M, presence of both unmethylated <strong>and</strong> methylated b<strong>and</strong>.Real-time quantitative gene expressionThe level of MAL mRNA expression <strong>in</strong> cell l<strong>in</strong>es (n = 46),primary colorectal carc<strong>in</strong>omas (n = 16), <strong>and</strong> <strong>no</strong>rmalmucosa (n = 3) was assessed by quantitative real timePCR. There was a strong association between MAL promoterhypermethylation <strong>and</strong> reduced or lost gene expressionamong cell l<strong>in</strong>es (P = 0.041; Figure 4). Furthermore,the gene expression of MAL was up-regulated <strong>in</strong> coloncancer cell l<strong>in</strong>es after promoter demethylation <strong>in</strong>duced bythe comb<strong>in</strong>ed treatment 5-aza-2'-deoxycytid<strong>in</strong>e <strong>and</strong> trichostat<strong>in</strong>A (Figure 5). Treatment with the deacetylase<strong>in</strong>hibitor trichostat<strong>in</strong> A alone did <strong>no</strong>t <strong>in</strong>crease MALexpression, whereas treatment with the DNA demethylat<strong>in</strong>g5-aza-2'-deoxycytid<strong>in</strong>e led to high expression <strong>in</strong> HT29cells, but more moderate levels <strong>in</strong> HCT15 cells (Figure 5).Among primary colorectal carc<strong>in</strong>omas, those harbour<strong>in</strong>gpromoter hypermethylation of MAL (n = 13) expressedsomewhat lower levels of MAL mRNA compared with theunmethylated tumours (n = 3), although <strong>no</strong>t statisticallysignificant (Figure 4).MAL prote<strong>in</strong> expression is lost <strong>in</strong> colorectal carc<strong>in</strong>omasTo evaluate the immu<strong>no</strong>histochemistry analyses of MAL,kidney <strong>and</strong> heart muscle tissues were <strong>in</strong>cluded as positive<strong>and</strong> negative controls, respectively (Figure 6A–B) [30].From the 231 scorable colorectal tissue cores, i.e. thoseconta<strong>in</strong><strong>in</strong>g malignant colorectal epithelial tissue, 198were negative for MAL sta<strong>in</strong><strong>in</strong>g (Figure 6C–D). Twentyn<strong>in</strong>eof these had positive sta<strong>in</strong><strong>in</strong>g <strong>in</strong> <strong>no</strong>n-epithelial tissuecomponents with<strong>in</strong> the same tissue cores, ma<strong>in</strong>ly <strong>in</strong> neurons<strong>and</strong> blood vessels (<strong>no</strong>t shown). In comparison, allthe sections of <strong>no</strong>rmal colon tissue conta<strong>in</strong>ed positivesta<strong>in</strong><strong>in</strong>g for MAL <strong>in</strong> the epithelial cells (Figure 6E–F).DiscussionIn the present study, we have demonstrated that asequence with<strong>in</strong> the MAL promoter close to the transcriptionstart is hypermethylated <strong>in</strong> the vast majority of malignant,as well as <strong>in</strong> benign colorectal tumours, <strong>in</strong> contrastto <strong>no</strong>rmal colon mucosa samples which are unmethylated,<strong>and</strong> we contend that MAL rema<strong>in</strong>s a promis<strong>in</strong>g diag<strong>no</strong>sticbiomarker for early colorectal tumorigenesis [12].The ade<strong>no</strong>mas <strong>and</strong> carc<strong>in</strong>omas analyzed <strong>in</strong> the presentstudy are from unselected cl<strong>in</strong>ical series <strong>and</strong> are thereforerepresentative for the average risk population. However,the equal distribution between MSI <strong>and</strong> MSS carc<strong>in</strong>omas<strong>in</strong> the present study is <strong>no</strong>t representative for a consecutiveseries.Hypermethylation of MAL has, by quantitative methylation-specificpolymerase cha<strong>in</strong> reaction (MSP), previouslybeen shown by others to be present only <strong>in</strong> a small fraction(6%, 2/34) of colon carc<strong>in</strong>omas [31], even thoughthe expression of MAL was reported to be reduced/lost <strong>in</strong>the majority of colorectal tumours [11,17,31]. In contrast,we report here a significantly higher methylation fre-Page 6 of 11(page number <strong>no</strong>t for citation purposes)