Introduction to Molecular Dynamics with GROMACS
Introduction to Molecular Dynamics with GROMACS
Introduction to Molecular Dynamics with GROMACS
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
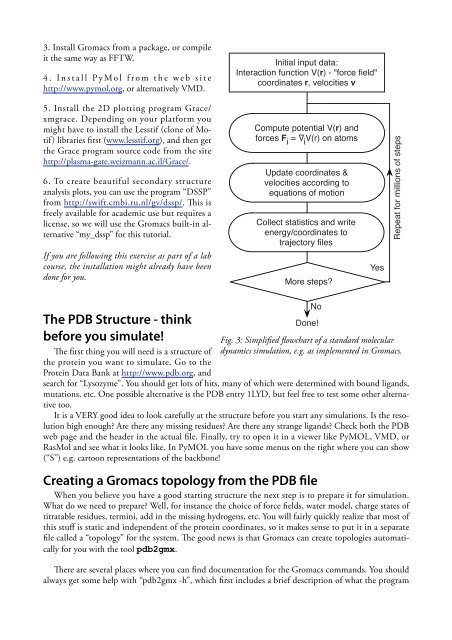
3. Install Gromacs from a package, or compileit the same way as FFTW.4 . I n s t a l l P y M o l f r o m t h e w e b s i t ehttp://www.pymol.org, or alternatively VMD.Initial input data:Interaction function V(r) - "force field"coordinates r, velocities v5. Install the 2D plotting program Grace/xmgrace. Depending on your platform youmight have <strong>to</strong> install the Lesstif (clone of Motif)libraries first (www.lesstif.org), and then getthe Grace program source code from the sitehttp://plasma-gate.weizmann.ac.il/Grace/.6. To create beautiful secondary structureanalysis plots, you can use the program “DSSP”from http://swift.cmbi.ru.nl/gv/dssp/. This isfreely available for academic use but requires alicense, so we will use the Gromacs built-in alternative“my_dssp” for this tu<strong>to</strong>rial.If you are following this exercise as part of a labcourse, the installation might already have beendone for you.Compute potential V(r) andforces F i=iV(r) on a<strong>to</strong>msUpdate coordinates &velocities according <strong>to</strong>equations of motionCollect statistics and writeenergy/coordinates <strong>to</strong>trajec<strong>to</strong>ry filesMore steps?YesRepeat for millions of stepsThe PDB Structure - thinkbefore you simulate!The first thing you will need is a structure ofthe protein you want <strong>to</strong> simulate. Go <strong>to</strong> theProtein Data Bank at http://www.pdb.org, andDone!search for “Lysozyme”. You should get lots of hits, many of which were determined <strong>with</strong> bound ligands,mutations, etc. One possible alternative is the PDB entry 1LYD, but feel free <strong>to</strong> test some other alternative<strong>to</strong>o.It is a VERY good idea <strong>to</strong> look carefully at the structure before you start any simulations. Is the resolutionhigh enough? Are there any missing residues? Are there any strange ligands? Check both the PDBweb page and the header in the actual file. Finally, try <strong>to</strong> open it in a viewer like PyMOL, VMD, orRasMol and see what it looks like. In PyMOL you have some menus on the right where you can show(“S”) e.g. car<strong>to</strong>on representations of the backbone!Creating a Gromacs <strong>to</strong>pology from the PDB fileWhen you believe you have a good starting structure the next step is <strong>to</strong> prepare it for simulation.What do we need <strong>to</strong> prepare? Well, for instance the choice of force fields, water model, charge states oftitratable residues, termini, add in the missing hydrogens, etc. You will fairly quickly realize that most ofthis stuff is static and independent of the protein coordinates, so it makes sense <strong>to</strong> put it in a separatefile called a “<strong>to</strong>pology” for the system. The good news is that Gromacs can create <strong>to</strong>pologies au<strong>to</strong>maticallyfor you <strong>with</strong> the <strong>to</strong>ol pdb2gmx.There are several places where you can find documentation for the Gromacs commands. You shouldalways get some help <strong>with</strong> “pdb2gmx -h”, which first includes a brief description of what the programNoFig. 3: Simplified flowchart of a standard moleculardynamics simulation, e.g. as implemented in Gromacs.















![[VAR]=Notes on variational calculus](https://img.yumpu.com/35639168/1/190x245/varnotes-on-variational-calculus.jpg?quality=85)