Introduction to Molecular Dynamics with GROMACS
Introduction to Molecular Dynamics with GROMACS
Introduction to Molecular Dynamics with GROMACS
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
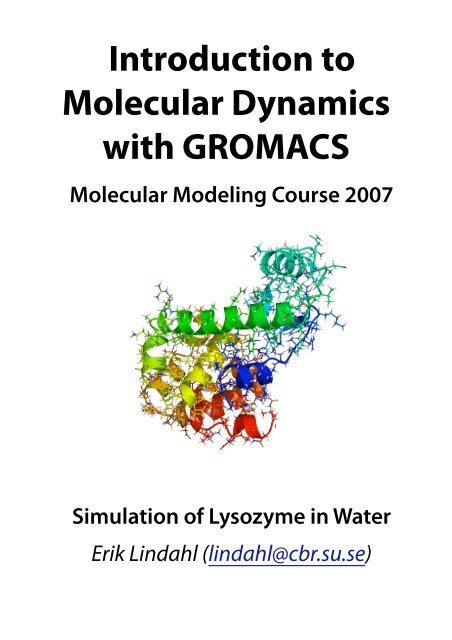
<strong>Introduction</strong> <strong>to</strong><strong>Molecular</strong> <strong>Dynamics</strong><strong>with</strong> <strong>GROMACS</strong><strong>Molecular</strong> Modeling Course 2007Simulation of Lysozyme in WaterErik Lindahl (lindahl@cbr.su.se)
Summary<strong>Molecular</strong> simulation is a very powerful <strong>to</strong>olbox in modern molecular modeling, and enables us <strong>to</strong>follow and understand structure and dynamics <strong>with</strong> extreme detail – literally on scales where motion ofindividual a<strong>to</strong>ms can be tracked. This task will focus on the two most commonly used methods, namelyenergy minimization and molecular dynamics that respectively optimize structure and simulate thenatural motion of biological macromolecules. we are first going <strong>to</strong> set up your Gromacs environments,have a look at the structure, prepare the input files necessary for simulation, solvate the structure in water,minimize & equilibrate it, and finally perform a short production simulation. After this we’ll testsome simple analysis programs that are part of Gromacs.You should write a brief report about your work and describe your findings. In particular, try <strong>to</strong>think about why you are using particular algorithms/choices, and whether there are alternatives.<strong>Molecular</strong> motions occur over a wide range of scales in both time and space, and the choice of approach<strong>to</strong> study them depends on the question asked. <strong>Molecular</strong> simulation is far from the only availablemethod, and when the aim e.g. is <strong>to</strong> predict the structure of a protein it is often more efficient <strong>to</strong>use bioinformatics instead of spending thousands or millions of CPU hours. Ideally, the timedependentSchrödinger equation should be able <strong>to</strong> predict all properties of any molecule <strong>with</strong> arbitraryprecision ab initio. However, as soon as more than a handful of particles are involved it is necessary <strong>to</strong>introduce approximations. For most biomolecular systems we therefore choose <strong>to</strong> work <strong>with</strong> empiricalparameterizations of models instead, for instance classical Coulomb interactions between pointlikea<strong>to</strong>mic charges rather than a quantum description of the electrons. These models are not only orders ofmagnitude faster, but since they have been parameterized from experiments they also perform betterwhen it comes <strong>to</strong> reproducing observations on microsecond scale (Fig. 1), rather than extrapolatingquantum models 10 orders of magnitude. The first molecular dynamics simulation was performed aslate as 1957, although it was not until the 1970’s that it was possible <strong>to</strong> simulate water and biomolecules.Background & TheoryMacroscopic properties measured in an experiment are not direct observations, but averages over billionsof molecules representing a statistical mechanics ensemble. This has deep theoretical implicationsthat are covered in great detail in the literature, but even from a practical point of view there are importantconsequences: (i) It is not sufficient <strong>to</strong> work <strong>with</strong> individual structures, but systems have <strong>to</strong> be expanded<strong>to</strong> generate a representative ensemble of structures at the given experimental conditions, e.g.temperature and pressure. (ii) Thermodynamic equilibrium properties related <strong>to</strong> free energy, such asbinding constant, solubilities, and relative stability cannot be calculated directly from individual simulations,but require more elaborate techniques. (iii) For equilibrium properties (in contrast <strong>to</strong> kinetic) theaim is <strong>to</strong> examine structure ensembles, and not necessarily <strong>to</strong> reproduce individual a<strong>to</strong>mic trajec<strong>to</strong>ries!The two most common ways <strong>to</strong> generate statistically faithful equilibrium ensembles are Monte Carloand <strong>Molecular</strong> <strong>Dynamics</strong> simulations. Monte Carlo simulations rely on designing intelligent moves <strong>to</strong>generate new conformations, but since these are fairly difficult <strong>to</strong> invent most simulations tend <strong>to</strong> doclassical dynamics <strong>with</strong> New<strong>to</strong>n’s equations of motion since this also has the advantage of accuratelyreproducing kinetics of non-equilibrium properties such as diffusion or folding times. When a startingconfiguration is very far from equilibrium, large forces can cause the simulation <strong>to</strong> crash or dis<strong>to</strong>rt thesystem, and for this reason it is usually necessary <strong>to</strong> start <strong>with</strong> an Energy Minimization of the systemprior <strong>to</strong> the molecular dynamics simulation. In addition, energy minimizations are commonly used <strong>to</strong>refine low-resolution experimental structures.All classical simulation methods rely on more or less empirical approximations called Force fields <strong>to</strong>calculate interactions and evaluate the potential energy of the system as a function of point-like a<strong>to</strong>miccoordinates. A force field consists of both the set of equations used <strong>to</strong> calculate the potential energy andforces from particle coordinates, as well as a collection of parameters used in the equations.
3. Install Gromacs from a package, or compileit the same way as FFTW.4 . I n s t a l l P y M o l f r o m t h e w e b s i t ehttp://www.pymol.org, or alternatively VMD.Initial input data:Interaction function V(r) - "force field"coordinates r, velocities v5. Install the 2D plotting program Grace/xmgrace. Depending on your platform youmight have <strong>to</strong> install the Lesstif (clone of Motif)libraries first (www.lesstif.org), and then getthe Grace program source code from the sitehttp://plasma-gate.weizmann.ac.il/Grace/.6. To create beautiful secondary structureanalysis plots, you can use the program “DSSP”from http://swift.cmbi.ru.nl/gv/dssp/. This isfreely available for academic use but requires alicense, so we will use the Gromacs built-in alternative“my_dssp” for this tu<strong>to</strong>rial.If you are following this exercise as part of a labcourse, the installation might already have beendone for you.Compute potential V(r) andforces F i=iV(r) on a<strong>to</strong>msUpdate coordinates &velocities according <strong>to</strong>equations of motionCollect statistics and writeenergy/coordinates <strong>to</strong>trajec<strong>to</strong>ry filesMore steps?YesRepeat for millions of stepsThe PDB Structure - thinkbefore you simulate!The first thing you will need is a structure ofthe protein you want <strong>to</strong> simulate. Go <strong>to</strong> theProtein Data Bank at http://www.pdb.org, andDone!search for “Lysozyme”. You should get lots of hits, many of which were determined <strong>with</strong> bound ligands,mutations, etc. One possible alternative is the PDB entry 1LYD, but feel free <strong>to</strong> test some other alternative<strong>to</strong>o.It is a VERY good idea <strong>to</strong> look carefully at the structure before you start any simulations. Is the resolutionhigh enough? Are there any missing residues? Are there any strange ligands? Check both the PDBweb page and the header in the actual file. Finally, try <strong>to</strong> open it in a viewer like PyMOL, VMD, orRasMol and see what it looks like. In PyMOL you have some menus on the right where you can show(“S”) e.g. car<strong>to</strong>on representations of the backbone!Creating a Gromacs <strong>to</strong>pology from the PDB fileWhen you believe you have a good starting structure the next step is <strong>to</strong> prepare it for simulation.What do we need <strong>to</strong> prepare? Well, for instance the choice of force fields, water model, charge states oftitratable residues, termini, add in the missing hydrogens, etc. You will fairly quickly realize that most ofthis stuff is static and independent of the protein coordinates, so it makes sense <strong>to</strong> put it in a separatefile called a “<strong>to</strong>pology” for the system. The good news is that Gromacs can create <strong>to</strong>pologies au<strong>to</strong>maticallyfor you <strong>with</strong> the <strong>to</strong>ol pdb2gmx.There are several places where you can find documentation for the Gromacs commands. You shouldalways get some help <strong>with</strong> “pdb2gmx -h”, which first includes a brief description of what the programNoFig. 3: Simplified flowchart of a standard moleculardynamics simulation, e.g. as implemented in Gromacs.
does, then a list of (optional) input/output files, and finally a list of available flags. The same informationis present <strong>with</strong> “man pdb2gmx” (if it was properly installed), and you can also use thewww.gromacs.org website: Choose “documentation” in the <strong>to</strong>p menu, and then “manuals” in the leftsidemenu. There is both an extensive book describing everything and a shorter online reference <strong>to</strong> thecommands and file formats.For the first command we’ll help you. pdb2gmx should read its input from a coordinate file, and justfor fun we’ll pick a specific water model and force field. Typepdb2gmx -f 1LYD.pdb -water tip3pThe program will ask you for a force field - for this tu<strong>to</strong>rial I suggest that you use “OPLS-AA/L”, andthen write out lots of information. The important thing <strong>to</strong> look for is if there were any errors or warnings(it will say at the end). Otherwise you are probably fine. Read the help text of pdb2gmx (-h) andtry <strong>to</strong> find out what the different files it just created do, and what options are available for pdb2gmx!The important output files are (find out yourself what they do)conf.gro <strong>to</strong>pol.<strong>to</strong>p posre.itpIf you issue the command several times, Gromacs will back up old files so their names start <strong>with</strong> ahash mark (#) - you can safely ignore (or erase) these for now.Adding solvent water around the proteinIf you started a simulation now it would be in vacuo. This was actually the norm 25-30 years ago,since it is much faster, but since we want this simulation <strong>to</strong> be accurate you are going <strong>to</strong> add solvent.However, before adding solvent we have <strong>to</strong> determine how big the simulation box should be, andwhat shape <strong>to</strong> use for it. There should be at least half a cu<strong>to</strong>ff between the molecule and the box side,and preferably more. Due <strong>to</strong> the limited time you be cheap here and only use 0.5nm margin betweenthe protein and box. To further reduce the volume of the box you could explore the box type optionsand for instance test a “rhombic dodecahedron” box instead of a cubic one. The program you should use<strong>to</strong> alter the box is “editconf”. This time you will have <strong>to</strong> figure out the exact command(s) yourself,but the primary options <strong>to</strong> look at in the documentation are “–f”, “–d”, “–o”, and “–bt”.As an exercise, you could try a couple of different box sizes (the volume is written on the output!)and also other box shapes like “cubic” or “octahedron”. What effect does it have on the volume?The last step before the simulation is <strong>to</strong> add water in the box <strong>to</strong> solvate the protein. This is done byusing a small pre-equilibrated system of water coordinates that is repeated over the box, and overlappingwater molecules removed. The Lysozyme system will require roughly 6000 water molecules <strong>with</strong> 0.5nm<strong>to</strong> the box side, which increases the number of a<strong>to</strong>ms significantly (from 2900 <strong>to</strong> over 20,000).<strong>GROMACS</strong> does not use a special pre-equilibrated system for TIP3P water since water coordinates canbe used <strong>with</strong> any model – the actual parameters are s<strong>to</strong>red in the <strong>to</strong>pology and force field. For this reasonthe program only provides pre-equilibrated coordinates for a small 216-water system using a watermodel called “SPC” (simple point charge). The best program <strong>to</strong> add solvent water is “genbox”, i.e.you will generate a box of solvent. You can select the SPC water coordinates <strong>to</strong> add <strong>with</strong> the flag “–csspc216.gro”, and for the rest of the choices you can use the documentation. Remember that your<strong>to</strong>pology needs <strong>to</strong> be updated <strong>with</strong> the new water molecules! The easiest way <strong>to</strong> do that is <strong>to</strong> supply the<strong>to</strong>pology <strong>to</strong> the genbox command.Before you continue it is once more a good idea <strong>to</strong> look at this system in PyMOL. However, Py-MOL and most other programs cannot read the Gromacs gro file directly. There are two simple ways <strong>to</strong>create a PDB file for PyMOL:
1. Use the ability of all Gromacs programs <strong>to</strong> write output in alternative formats, e.g.:genbox –cp box.gro –cs spc216.gro –p <strong>to</strong>pol.<strong>to</strong>p –o solvated.pdb2. Use the Gromacs editconf program <strong>to</strong> convert it (use -h <strong>to</strong> get help on the options):editconf -f solvated.gro -o solvated.pdbIf you just use the commands like this, the resulting structure might look a bit strange, <strong>with</strong> water ina rectangular box even if the system is triclinic/octahedron/dodecahedron. This is actually fine, and dependson the algorithms used <strong>to</strong> add the water combined <strong>with</strong> periodic boundary conditions. However,<strong>to</strong> get a beautiful-looking unit cell you can try the commands (backslash means the entire commandshould be on a single line):trjconv -s solvated.gro -f solvated.gro -o solv_triclinic.pdb \-pbc inbox -ur trictrjconv -s solvated.gro -f solvated.gro -o solv_compact.pdb \-pbc inbox -ur compactWe’re not going <strong>to</strong> tell you what they do - play around and use the -h option <strong>to</strong> learn more :-)Running energy minimizationThe added hydrogens and broken hydrogen bond network in water would lead <strong>to</strong> quite large forcesand structure dis<strong>to</strong>rtion if molecular dynamics was started immediately. To remove these forces it is necessary<strong>to</strong> first run a short energy minimization. The aim is not <strong>to</strong> reach any local energy minimum, soe.g. 200 <strong>to</strong> 500 steps of steepest descent works very well as a stable rather than maximally efficient minimization.Nonbonded interactions and other settings are specified in a parameter file (em.mdp); it isonly necessary <strong>to</strong> specify parameters where we deviate from the default value, for example:------em.mdp------integra<strong>to</strong>r = steepnsteps = 200nstlist = 10rlist = 1.0coulombtype = pmercoulomb = 1.0vdw-type = cut-offrvdw = 1.0nstenergy = 10------------------Comments on the parameters used:We choose a standard cut-off of 1.0 nm, both for the neighborlist generation and the coulomb andLennard-Jones interactions. nstlist=10 means it is updated at least every 10 steps, but for energy minimizationit will usually be every step. Energies and other statistical data are s<strong>to</strong>red every 10 steps (nstenergy),and we have chosen the more expensive Particle-Mesh-Ewald summation for electrostatic interactions. Thetreatment of nonbonded interactions is frequently bordering <strong>to</strong> religion. One camp advocates standardcu<strong>to</strong>ffs are fine, another swears by switched-off interactions, while the third wouldn’t even consider anythingbut PME. One argument in this context is that ‘true’ interactions should conserve energy, which isviolated by sharp cut-offs since the force is no longer the exact derivative of the potential. On the otherhand, just because an interaction conserves energy doesn’t mean it describes nature accurately. In practice,the difference is most pronounced for systems that are very small or <strong>with</strong> large charges, but the key lesson is
eally that it is a trade-off. PME is great, but also clearly slower than cut-offs. Longer cut-offs are alwaysbetter than short ones (but slower), and while switched interactions improve energy conservation they introduceartificially large forces. Using PME is the safe option, but if that’s not fast enough it is worth investigatingreaction-field or cut-off interactions. It is also a good idea <strong>to</strong> check and follow the recommendedsettings for the force field used.<strong>GROMACS</strong> uses a separate preprocessing program grompp <strong>to</strong> collect parameters, <strong>to</strong>pology, andcoordinates in<strong>to</strong> a single run input file (e.g. em.tpr) from which the simulation is then started (thismakes it easier <strong>to</strong> prepare a run on your workstation but run it on a separate supercomputer).As we just said, the command grompp prepares run input files. You will need <strong>to</strong> provide it <strong>with</strong>the parameter file above, the <strong>to</strong>pology, a set of coordinates, and give the run input file some name (otherwiseit will get the default <strong>to</strong>pol.tpr name). The program <strong>to</strong> actually run the energy minimization ismdrun - this is the very heart of Gromacs. mdrun has a really smart shortcut flag called “-deffnm” <strong>to</strong> use a string you provide (e.g. “em”) as the default name for all options. If you want <strong>to</strong>get some status update during the run it is also smart <strong>to</strong> turn on the verbosity flag (guess the letter,or consult the documentation). The minimization should finish in a couple of minutes.Carefully equilibrating the water around the proteinTo avoid unnecessary dis<strong>to</strong>rtion of the protein when the molecular dynamics simulation is started,we first perform an equilibration run where all heavy protein a<strong>to</strong>ms are restrained <strong>to</strong> their starting positions(using the file posre.itp generated earlier) while the water is relaxing around the structure. Thisshould really be 50-100ps, but if that appears <strong>to</strong> be <strong>to</strong>o slow you can make do <strong>with</strong> 10ps (5000 steps)here. Bonds will be constrained <strong>to</strong> enable 2 fs time steps. Other settings are identical <strong>to</strong> energy minimization,but for molecular dynamics we also control the temperature and pressure <strong>with</strong> the Berendsenweak coupling algorithm. This is mostly done by extending the parameter file used above:------pr.mdp------integra<strong>to</strong>r = mdnsteps = 5000dt = 0.002nstlist = 10rlist = 1.0coulombtype = pmercoulomb = 1.0vdw-type = cut-offrvdw = 1.0tcoupl= Berendsentc-grps= protein non-proteintau-t = 0.1 0.1ref-t = 298 298Pcoupl= Berendsentau-p = 1.0compressibility = 5e-5 5e-5 5e-5 0 0 0ref-p = 1.0nstenergy = 100define= -DPOSRES
------------------For a small protein like Lysozyme it should be more than enough <strong>with</strong> 100 ps (50,000 steps) for thewater <strong>to</strong> fully equilibrate around it, but in a large membrane system the slow lipid motions can requireseveral nanoseconds of relaxation. The only way <strong>to</strong> know for certain is <strong>to</strong> watch the potential energy,and extend the equilibration until it has converged. To run this equilibration in <strong>GROMACS</strong> youshould once more use the programs grompp and mdrun, but (i) apply the new parameters, and (ii)start from the final coordinates of the energy minimization.Running the production simulationThe difference between equilibration and production run will be minimal: the position restraints andpressure coupling are turned off, we decide how often <strong>to</strong> write output coordinates <strong>to</strong> analyze (say, every100 steps), and start a significantly longer simulation. How long depends on what you are studying, andthat should be decided before starting any simulations! For decent sampling the simulation should be atleast 10 times longer than the phenomena you are studying, which unfortunately sometimes conflicts<strong>with</strong> reality and available computer resources. This will depend a lot on the performance of your computer,but one option is <strong>to</strong> first run a very short simulation so you have something <strong>to</strong> work <strong>with</strong> whilelearning the analysis <strong>to</strong>ols, and in the meantime you can run a longer simulation in the background.Select the number of steps and output frequencies yourself, but for reasons you’ll see later it is recommended<strong>to</strong> write coordinates at least every picosecond, i.e. 500 steps.------run.mdp------integra<strong>to</strong>r = mdnsteps = < select yourself! >dt = 0.002nstlist = 10rlist = 1.0coulombtype = pmercoulomb = 1.0vdw-type = cut-offrvdw = 1.0tcoupl= Berendsentc-grps= protein non-proteintau-t = 0.1 0.1ref-t = 298 298nstxout = < select yourself! >nstvout = < select yourself! >nstxtcout = < select yourself! >nstenergy = < select yourself! >------------------S<strong>to</strong>ring full precision coordinates/velocities (nstxout/nstvout) enables restart if runs crash (poweroutage, full disk, etc.). The analysis normally only uses the compressed coordinates (nstxtcout). Use thedocumentation <strong>to</strong> figure out the names of the corresponding files. Perform the production run <strong>with</strong> thecommands grompp and mdrun (remember the verbosity flag <strong>to</strong> see how the run is progressing).Analysis of the simulation
You don’t have <strong>to</strong> do exactly these analyses, but try one or a couple of them that sound interestingand then use the documentation website or Google <strong>to</strong> see what else you can do if you prefer that. Atleast for the longer ones you will get clearer results <strong>with</strong> a longer trajec<strong>to</strong>ry.Deviation from X-ray structureOne of the most important fundamental properties <strong>to</strong> analyze is whether the protein is stable andclose <strong>to</strong> the experimental structure. The standard way <strong>to</strong> measure this is the root-mean-square displacement(RMSD) of all heavy a<strong>to</strong>ms <strong>with</strong> respect <strong>to</strong> the X-ray structure. <strong>GROMACS</strong> has a finished program<strong>to</strong> do this, called g_rms that takes a run input file and a trajec<strong>to</strong>ry <strong>to</strong> produce RMSD output.It is usually a good idea <strong>to</strong> use a reference structure (the tpr file) from the input before energy minimization,since that is the experimental one. The program will prompt both for a fit group, and thegroup <strong>to</strong> calculate RMSD for – choose “Protein-H” (protein except hydrogens) for both. The outputwill be written <strong>to</strong> rmsd.xvg, and if you installed the Grace program you will directly get a finished graphthat you can display <strong>with</strong> xmgrace rmsd.xvg.The RMSD increases pretty rapidly in the first part of the simulation, but stabilizes around 0.l9 nm,roughly the resolution of the X-ray structure. The difference is partly caused by limitations in the forcefield, but also because a<strong>to</strong>ms in the simulation are moving and vibrating around an equilibrium structure.A better measure can be obtained by first creating a running average structure from the simulationand comparing the running average <strong>to</strong> the X-ray structure, which would give a more realistic RMSDaround 0.16 nm.Comparing fluctuations <strong>with</strong> temperature fac<strong>to</strong>rsVibrations around the equilibrium are not random, but depend on local structure flexibility. Theroot-mean-square-fluctuation (RMSF) of each residue is straightforward <strong>to</strong> calculate over the trajec<strong>to</strong>ry,but more important they can be converted <strong>to</strong> temperature fac<strong>to</strong>rs that are also present for each a<strong>to</strong>m in aPDB file. Once again there is a program that will do the entire job - g_rmsf. The output can be written<strong>to</strong> an xvg file you can view <strong>with</strong> xmgrace, but also as temperature fac<strong>to</strong>rs directly in a PDB file(check the output file options for the program). When prompted, you can use the group “C-alpha” <strong>to</strong>get one value per residue. The overall agreement should be quite good (check the temperature fac<strong>to</strong>rfield in this and the input PDB file), which hopefully lends further credibility <strong>to</strong> the accuracy and stabilityof the simulation - your fluctuations are similar <strong>to</strong> those seen in the X-ray experiment.Secondary structure (Requires DSSP)Another measure of stability is the protein secondary structure. This can be calculated for each frame<strong>with</strong> a program such as DSSP. If the DSSP program is installed and the environment variable DSSPpoints <strong>to</strong> the binary (do_dssp -h will give you help on that), the <strong>GROMACS</strong> program do_dsspcan create time-resolved secondary structure plots. Since the program writes output in a special xpm (Xpixmap) format you probably also need the <strong>GROMACS</strong> program xpm2ps <strong>to</strong> convert it <strong>to</strong> postscript.Both commands are reasonably well documented, and the end result will be an eps file that you canview e.g. <strong>with</strong> ghostview or some other standard previewer application.Use the group “protein” for the secondary structure calculation. The DSSP secondary structure definitionis pretty tight, so it is quite normal for residues <strong>to</strong> fluctuate around the well-defined state, in particularat the ends of helices or sheets. For a (long) protein folding simulation, a DSSP plot would showhow the secondary structures form during the simulation. If you do not have DSSP installed, you cancompile the Gromacs built-in replacement “my_dssp” from the src/contrib direc<strong>to</strong>ry, or hope thatyour system administra<strong>to</strong>r already did so for you.
Distance and hydrogen bondsWith basic properties accurately reproduced, we can use the simulation <strong>to</strong> analyze more specific details.As an example, Lysozyme appears <strong>to</strong> be stabilized by hydrogen bonds between the residues GLU22and ARG137 (the numbers might be slightly different for structures other than 1LYD), so how muchdoes this fluctuate in the simulation, and are the hydrogen bonds intact? Gromacs has some very powerful<strong>to</strong>ols <strong>to</strong> analyze distances and hydrogen bonds, but <strong>to</strong> specify what <strong>to</strong> analyze you need an “indexfile” <strong>with</strong> the groups of a<strong>to</strong>ms in question. To create such a file, use the commandmake_ndx –f run.groAt the prompt, create a group for GLU22 <strong>with</strong> “r 22”, ARG137 <strong>with</strong> “r 137” (for residue 22 and137), and then “q” <strong>to</strong> save an index file as index.ndx. The distance and number of hydrogen bonds cannow be calculated <strong>with</strong> the commands g_dist and g_hbond. Both use more or less the same input(including the index file you just created). For the first program you might e.g. want the distance as afunction of time as output, and for the second the <strong>to</strong>tal number of hydrogen bonds vs. time. In bothcases you should select the two groups you just created. Two hydrogen bonds should be present almostthroughout the simulation, at least for 1LYD.Making a movieA normal movie uses roughly 30 frames/second, so a 10-second movie requires 300 simulation trajec<strong>to</strong>ryframes. To make a smooth movie the frames should not be more 1-2 ps apart, or it will just appear<strong>to</strong> shake nervously. To create a PDB-format trajec<strong>to</strong>ry (which can be read by PyMOL and VMD)<strong>with</strong> roughly 300 frames you can use the command trjconv. The option -e will set an end time (<strong>to</strong>get fewer frames), or you can use -skip e.g. <strong>to</strong> only use every fifth frame. Write the output in PDBformat by using .pdb as extension, e.g. movie.pdb.Choose the protein group for output rather than the entire system (water move <strong>to</strong>o fast). If you openthis trajec<strong>to</strong>ry in PyMOL you can immediately play it using the VCR-style controls on the bot<strong>to</strong>mright, adjust visual settings in the menus, and even use pho<strong>to</strong>realistic ray-tracing for all images in themovie. With MacPyMOL you can directly save the movie as a quicktime file, and on Linux you cansave it as a sequence of PNG images for assembly in another program. Rendering a movie only takes afew minutes, and if that doesn’t work you can still export PNG files. PyMOL can also create (rendered)PNG files <strong>with</strong> transparent background if you do: set ray_opaque_background, offConclusions & ReportThe main point of this exercise was <strong>to</strong> get your feet wet, and at least an idea of how <strong>to</strong> use MD inpractice. There are more advanced analysis <strong>to</strong>ols available, not <strong>to</strong> mention a wealth of simulation options.Write a brief report about your work and describe your findings. In particular, try <strong>to</strong> think aboutwhy you are using particular algorithms/choices, and whether there are alternatives!If you want <strong>to</strong> continue we’d like <strong>to</strong> recommend the Gromacs manual which is available from thewebsite: It goes quite a bit beyond just being a program manual, and explains many of the algorithmsand equations in detail! There are also a few videos & slides available online - Google is your friend!















![[VAR]=Notes on variational calculus](https://img.yumpu.com/35639168/1/190x245/varnotes-on-variational-calculus.jpg?quality=85)