10 A niversary of IIMCB
10 A niversary of IIMCB
10 A niversary of IIMCB
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
cross-linking, mass spectrometry, circular dichroism,<br />
limited proteolysis, etc.<br />
3. Protein engineering to obtain enzymes with new,<br />
useful features, in particular, altered substrate specificity<br />
(e.g. restriction enzymes that recognize and cut new<br />
sequences). Other protein engineering projects include<br />
attempts to design and obtain proteins with altered<br />
tertiary and quaternary structures.<br />
The research in all three sections is tightly integrated,<br />
as demonstrated by publications <strong>of</strong> articles comprising<br />
the combination <strong>of</strong> theoretical and experimental<br />
analyses, e.g. prediction and characterization <strong>of</strong> new RNA<br />
methyltransferases. In particular, protein engineering<br />
involves iterative protein structure model building, modelbased<br />
experiment planning, series <strong>of</strong> experimental analyses,<br />
and experiment-based improvement <strong>of</strong> the models and the<br />
tools used for model building.<br />
Recent highlights:<br />
Protein Structure Prediction<br />
The GeneSilico human predictors’ group and several<br />
servers developed in our laboratory achieved high positions<br />
in rankings <strong>of</strong> the 8 th Community-Wide Experiment on the<br />
Critical Assessment <strong>of</strong> Techniques for Protein Structure<br />
Prediction (CASP8), organized by the Protein Structure<br />
Prediction Center. Servers for protein structure prediction<br />
and model quality assessment achieved respectable<br />
positions in their respective categories. The most successful<br />
was the GeneSilico metaserver for protein disorder<br />
prediction, which ranked as number 1 in its category.<br />
These successful servers are freely available to all academic<br />
researchers via the GeneSilico toolkit.<br />
New rRNA modification enzymes predicted and<br />
confirmed experimentally<br />
Researchers from our laboratory predicted (with<br />
bioinformatics) and then experimentally confirmed that<br />
two so far uncharacterized open reading frames encode<br />
methyltransferases acting on 23S rRNA. YbeA is the m3Psi<br />
methyltransferase RlmH that targets nucleotide 1915,<br />
while YccW is the m5C methyltransferase RlmI specific for<br />
nucleotide 1962. Computational docking analysis revealed<br />
the potential way how RlmH may interact with the ribosome<br />
(as visualized on the figure p. 26). The experimental analysis<br />
was done in collaboration with a group in Odense headed by<br />
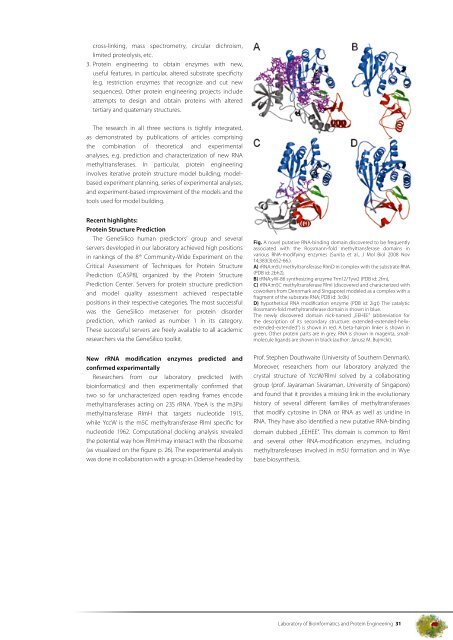
Fig. A novel putative RNA-binding domain discovered to be frequently<br />
associated with the Rossmann-fold methyltransferase domains in<br />
various RNA-modifying enzymes (Sunita et al., J Mol Biol 2008 Nov<br />
14;383(3):652-66.).<br />
A) rRNA:m5U methyltransferase RlmD in complex with the substrate RNA<br />
(PDB id: 2bh2),<br />
B) tRNA:yW-86 synthesizing enzyme Trm12/Tyw2 (PDB id: 2frn),<br />
C) rRNA:m5C methyltransferase RlmI (discovered and characterized with<br />
coworkers from Dennmark and Singapore) modeled as a complex with a<br />
fragment <strong>of</strong> the substrate RNA; PDB id: 3c0k)<br />
D) hypothetical RNA modification enzyme (PDB id: 2igt) The catalytic<br />
Rossmann-fold methyltransferase domain is shown in blue.<br />
The newly discovered domain nick-named „EEHEE” (abbreviation for<br />
the description <strong>of</strong> its secondary structure: extended-extended-helixextended-extended”)<br />
is shown in red. A beta-hairpin linker is shown in<br />
green. Other protein parts are in grey. RNA is shown in magenta, smallmolecule<br />
ligands are shown in black (author: Janusz M. Bujnicki).<br />
Pr<strong>of</strong>. Stephen Douthwaite (University <strong>of</strong> Southern Denmark).<br />
Moreover, researchers from our laboratory analyzed the<br />
crystal structure <strong>of</strong> YccW/RlmI solved by a collaborating<br />
group (pr<strong>of</strong>. Jayaraman Sivaraman, University <strong>of</strong> Singapore)<br />
and found that it provides a missing link in the evolutionary<br />
history <strong>of</strong> several different families <strong>of</strong> methyltransferases<br />
that modify cytosine in DNA or RNA as well as uridine in<br />
RNA. They have also identified a new putative RNA-binding<br />
domain dubbed „EEHEE”. This domain is common to RlmI<br />
and several other RNA-modification enzymes, including<br />
methyltransferases involved in m5U formation and in Wye<br />
base biosynthesis.<br />
Laboratory <strong>of</strong> Bioinformatics and Protein Engineering 31