Dynamic Programing
Dynamic Programing Dynamic Programing
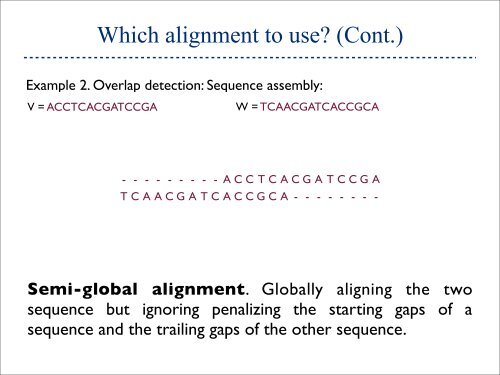
Which alignment to use (Cont.) Example 2. Overlap detection: Sequence assembly: V = ACCTCACGATCCGA W = TCAACGATCACCGCA - - - - - - - - - A C C T C A C G A T C C G A T C A A C G A T C A C C G C A - - - - - - - - Semi-global alignment. Globally aligning the two sequence but ignoring penalizing the starting gaps of a sequence and the trailing gaps of the other sequence.
Semi-Global Alignment finds optimal alignment without penalizing gaps on the ends of the alignment How to perform semi-global alignment Modify the basic Needleman-Wunsch algorithm: Set the first row and first column of the DP matrix to 0. v1 0 : 0 vi 0 : 0 vn 0 w1 ... wj ... wm 0 0 0 0 0 0 (I) (II) (III) +s(vi, wj) +gap +gap Optimal alignment score = max ( rown, columnm) Traceback starts at entry containing the optimal alignment score and ends at the first row or the first column.
- Page 3 and 4: Importance of Sequence Alignment
- Page 5 and 6: Alignment Operation Transforming on
- Page 7 and 8: Difficulties in measuring sequence
- Page 9 and 10: Efficient way to find a best alignm
- Page 11 and 12: Problems Solvable by Dynamic Progra
- Page 13: Property of DP problems • Have ov
- Page 16 and 17: I. Global Alignment Classes of Pair
- Page 18 and 19: Classes of Pairwise Alignment: I. G
- Page 20 and 21: Classes of Pairwise Alignment: I. G
- Page 22 and 23: Global Alignment spans all the resi
- Page 24 and 25: Scoring matrix represents a specifi
- Page 26 and 27: Needleman-Wunsch Algorithm (Cont.)
- Page 28 and 29: Needleman-Wunsch Algorithm (Cont.)
- Page 30 and 31: Needleman-Wunsch Algorithm (Cont.)
- Page 32 and 33: Needleman-Wunsch Algorithm Efficien
- Page 34 and 35: Needleman-Wunsch Algorithm for any
- Page 36 and 37: Needleman-Wunsch Algorithm for any
- Page 38 and 39: Local Alignment finds the most simi
- Page 40 and 41: Smith-Waterman Algorithm (Cont.)
- Page 42 and 43: Smith-Waterman Algorithm (Cont.) v1
- Page 44 and 45: Smith-Waterman Algorithm (Cont.) Ex
- Page 46 and 47: Which alignment to use Example 1. O
- Page 48 and 49: Which alignment to use Example 1. O
- Page 50 and 51: Which alignment to use Example 1. O
- Page 52 and 53: Which alignment to use example 1 co
- Page 56 and 57: Versatility of DP Algorithm • Mem
- Page 58 and 59: References - Gusfield D. Algorithms
Which alignment to use (Cont.)<br />
Example 2. Overlap detection: Sequence assembly:<br />
V = ACCTCACGATCCGA<br />
W = TCAACGATCACCGCA<br />
- - - - - - - - - A C C T C A C G A T C C G A<br />
T C A A C G A T C A C C G C A - - - - - - - -<br />
Semi-global alignment. Globally aligning the two<br />
sequence but ignoring penalizing the starting gaps of a<br />
sequence and the trailing gaps of the other sequence.