PYRUVATE KINASE - Toyobo
PYRUVATE KINASE - Toyobo
PYRUVATE KINASE - Toyobo
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
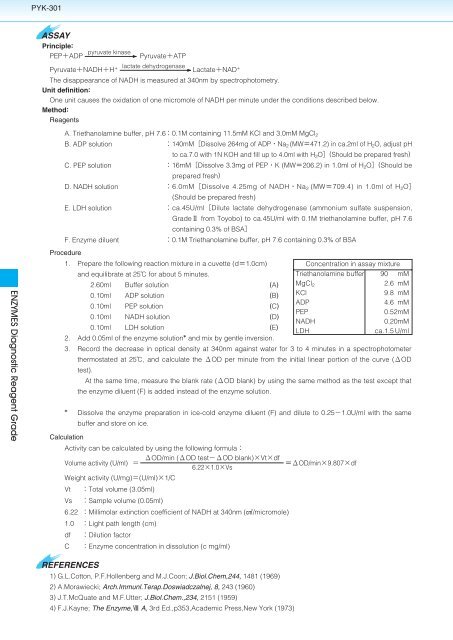
PYK-301<br />
ASSAY<br />
Principle:<br />
PEPADP<br />
pyruvate kinase<br />
PyruvateNADHH <br />
PyruvateATP<br />
lactate dehydrogenase<br />
LactateNAD <br />
The disappearance of NADH is measured at 340nm by spectrophotometry.<br />
Unit definition:<br />
One unit causes the oxidation of one micromole of NADH per minute under the conditions described below.<br />
Method:<br />
Reagents<br />
A. Triethanolamine buffer, pH 7.60.1M containing 11.5mM KCl and 3.0mM MgCl 2<br />
B. ADP solution<br />
140mMDissolve 264mg of ADPNa 2 (MW471.2) in ca.2ml of H 2 O, adjust pH<br />
to ca.7.0 with 1N KOH and fill up to 4.0ml with H 2 O(Should be prepared fresh)<br />
C. PEP solution<br />
16mMDissolve 3.3mg of PEPK (MW206.2) in 1.0ml of H 2 O(Should be<br />
prepared fresh)<br />
D. NADH solution<br />
6.0mMDissolve 4.25mg of NADHNa 2 (MW709.4) in 1.0ml of H 2 O<br />
(Should be prepared fresh)<br />
E. LDH solution<br />
ca.45U/mlDilute lactate dehydrogenase (ammonium sulfate suspension,<br />
Grade from <strong>Toyobo</strong>) to ca.45U/ml with 0.1M triethanolamine buffer, pH 7.6<br />
containing 0.3% of BSA<br />
F. Enzyme diluent<br />
0.1M Triethanolamine buffer, pH 7.6 containing 0.3% of BSA<br />
Procedure<br />
1. Prepare the following reaction mixture in a cuvette (d1.0cm)<br />
Concentration in assay mixture<br />
and equilibrate at 25 for about 5 minutes.<br />
Triethanolamine buffer 90 mM<br />
2.60ml Buffer solution (A) MgCl 2<br />
2.6 mM<br />
0.10ml ADP solution (B) KCl<br />
9.8 mM<br />
ADP<br />
4.6 mM<br />
0.10ml PEP solution (C)<br />
PEP<br />
0.52mM<br />
0.10ml NADH solution (D)<br />
NADH<br />
0.20mM<br />
0.10ml LDH solution (E)<br />
LDH<br />
ca.1.5 U/ml<br />
2. Add 0.05ml of the enzyme solution and mix by gentle inversion.<br />
3. Record the decrease in optical density at 340nm against water for 3 to 4 minutes in a spectrophotometer<br />
thermostated at 25, and calculate the OD per minute from the initial linear portion of the curve (OD<br />
test).<br />
At the same time, measure the blank rate (OD blank) by using the same method as the test except that<br />
the enzyme diluent (F) is added instead of the enzyme solution.<br />
<br />
Dissolve the enzyme preparation in ice-cold enzyme diluent (F) and dilute to 0.251.0U/ml with the same<br />
buffer and store on ice.<br />
Calculation<br />
Activity can be calculated by using the following formula<br />
OD/min (OD testOD blank)Vtdf<br />
Volume activity (U/ml) <br />
6.221.0Vs<br />
Weight activity (U/mg)(U/ml)1/C<br />
OD/min9.807df<br />
Vt<br />
Vs<br />
Total volume (3.05ml)<br />
Sample volume (0.05ml)<br />
6.22 Millimolar extinction coefficient of NADH at 340nm (F/micromole)<br />
1.0 Light path length (cm)<br />
df<br />
C<br />
Dilution factor<br />
Enzyme concentration in dissolution (c mg/ml)<br />
REFERENCES<br />
1) G.L.Cotton, P.F.Hollenberg and M.J.Coon; J.Biol.Chem,244, 1481 (1969)<br />
2) A.Morawiecki; Arch.Immunl.Terap.Doswiadczalnej, 8, 243 (1960)<br />
3) J.T.McQuate and M.F.Utter; J.Biol.Chem.,234, 2151 (1959)<br />
4) F.J.Kayne; The Enzyme, A, 3rd Ed.,p353,Academic Press,New York (1973)