Elektronika 2009-11.pdf - Instytut Systemów Elektronicznych
Elektronika 2009-11.pdf - Instytut Systemów Elektronicznych
Elektronika 2009-11.pdf - Instytut Systemów Elektronicznych
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Medical pattern intelligent recognition based<br />
on linguistic modelling of 3D coronary vessels<br />
visualisations<br />
(Lingwistyczne modelowanie przestrzennych rekonstrukcji unaczynienia<br />
wieńcowego w inteligentnym rozpoznawaniu zmian patologicznych)<br />
mgr inż. MIROSŁAW TRZUPEK, prof. dr hab. MAREK R. OGIELA,<br />
prof. dr hab. inż. RYSZARD TADEUSIEWICZ<br />
AGH University of Science and Technology, Institute of Automatics, Bio-Cybernetics Laboratory, Krakow<br />
Characteristic for the coronary vessels in different patients with<br />
various pathologic forms observed on flat, i.e. 2D, images is a<br />
certain level of repetitiveness and regularity. The same images<br />
registered by a machine rendering 3D images (helical CT<br />
scanner) feature a much greater number of visible details,<br />
which, however, results also in the increase of the number of<br />
both individual differences and those between various examinations<br />
of the same patient. Having considering these difficulties,<br />
a decision was made to apply the methods of automatic<br />
image understanding for the interpretation of the images considered,<br />
which consequently leads to their semantic descriptions.<br />
Thanks to such posing of the problem, the diagnosis put<br />
forth (being naturally but a suggestion for the physician making<br />
the actual decision) will account for a greater number of<br />
factors and use better the available medical information.<br />
Additionally, it is worth pointing out that - even though this<br />
is not the main focus of the work presented here - that the solution<br />
of problems related to the automatic production of intelligent<br />
(cognitive) descriptions of the 3D medical images in<br />
question may be a significant contribution to solving at least<br />
some of the problems connected to the smart archiving of this<br />
type of data and the finding of semantic image data meeting<br />
the semantic criteria delivered through sample image patterns<br />
in medical multimedia databases.<br />
The goal of research and description<br />
of the problem<br />
Problems of modelling biomedical structure shapes are extremely<br />
interesting from the scientific point of view, but also in<br />
connection with the development of medical diagnostic support<br />
systems. They concern various structures diagnosed<br />
using various modalities. However, in such research, we extremely<br />
frequently see the use of an approach based on neutral<br />
networks for shape modelling. For instance, such models<br />
can be built using Kohonen networks [1]. However, it is worth<br />
noting that such solutions are usually limited by the network<br />
topology, which, although it allows you to operate on vectors<br />
of features informative for the objects researched, does not<br />
allow the image to be analysed or the organ to be reconstructed<br />
in a holistic fashion, i.e. taking into account the full<br />
dimensions of the image [2-4]. This only becomes possible<br />
when we create linguistic descriptions using image grammars<br />
based on graph formalisms. By definition, such grammars<br />
have been dedicated for analysing image scenes, which in the<br />
case of medical visualisation means that they can be used to<br />
model multi-object structures or to conduct the meaning analysis<br />
of spatial reconstructions of heart tomograms, which will be<br />
demonstrated later in this publication.<br />
Graph-based semantic models<br />
of the heart’s coronary vessels<br />
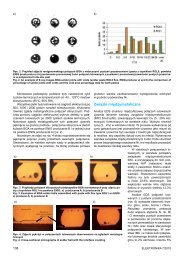
In order to analyze a 3D reconstruction (visualisations for various<br />
patients obtained during diagnostic examinations of the<br />
heart with a helical CT scanner with 64 detectors), it becomes<br />
necessary to select the appropriate projection showing lesions<br />
in vessels in a way that enables them to be analysed on<br />
a plane. In our research we have attempted to automate the<br />
procedure of finding such a projection by using selected geometric<br />
transformations during image processing. Next, to enable<br />
a linguistic representation of the spatial reconstructions<br />
studied, the coronary vessels shown in them had been subjected<br />
to the operation of thinning, referred to as skeletonizing.<br />
This operation allows us to obtain a skeleton of the arteries<br />
under consideration with the thickness of one unit. This skeleton<br />
can then be subjected to the operation of labelling, which<br />
determines the start and end points of main and surrounding<br />
branches of coronary arteries in it. These points will constitute<br />
the peaks of a graph modelling the spatial structure of the<br />
coronary vessels of the heart.<br />
The next step is labeling them by giving each located informative<br />
point the appropriate label from the set of peak labels<br />
which unambiguously identify individual coronary arteries<br />
forming parts of the structure analysed. For the left coronary<br />
artery they have been defined as follows: LCA - left coronary<br />
artery, LAD - anterior interventricular branch (left anterior descending),<br />
CX - circumflex branch, L - lateral branch, LM - left<br />
marginal branch. And for the right coronary artery they have<br />
been defined as follows: RCA - right coronary artery, A - atrial<br />
branch, RM - right marginal branch, PI - posterior interventricular<br />
branch, RP - right posterolateral branch. This way, all<br />
initial and final points of coronary vessels as well as all points<br />
where main vessels branch or change into lower level vessels<br />
have been determined and labelled as appropriate. After this<br />
operation, the coronary vascularization tree is divided into<br />
sections which constitute the edges of a graph modelling the<br />
examined coronary arteries. Mutual spatial relations that may<br />
occur between elements of the vascular structure represented<br />
by a graph are described by the set of edge labels. The elements<br />
of this set have been defined by introducing the appropriate<br />
spatial relations; vertical - defined by the set of labels<br />
α, β,…, µ and horizontal - defined by the set of labels 1, 2,…,<br />
24 on a hypothetical sphere surrounding the heart muscle.<br />
These labels designate individual final intervals, each of which<br />
has the angular spread of 15°. Then, depending on the location,<br />
terminal edge labels are assigned to all branches identified<br />
by the beginnings and ends of the appropriate sections of<br />
coronary arteries.<br />
ELEKTRONIKA 11/<strong>2009</strong> 9