On the Analysis of Optical Mapping Data - University of Wisconsin ...
On the Analysis of Optical Mapping Data - University of Wisconsin ...
On the Analysis of Optical Mapping Data - University of Wisconsin ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
34<br />
250<br />
200<br />
0.700 − 0.005<br />
0 50 100 150 200 250<br />
0.750 − 0.005 0.800 − 0.005<br />
150<br />
100<br />
50<br />
Quantiles <strong>of</strong> GMO7535 fragment lengths<br />
0<br />
250<br />
0.700 − 0.003 0.750 − 0.003<br />
0.700 − 0.001 0.750 − 0.001<br />
0.800 − 0.003<br />
0.800 − 0.001<br />
250<br />
200<br />
150<br />
100<br />
50<br />
0<br />
200<br />
150<br />
100<br />
50<br />
0<br />
0 50 100 150 200 250<br />
Quantiles <strong>of</strong> fragment lengths in simulated data<br />
0 50 100 150 200 250<br />
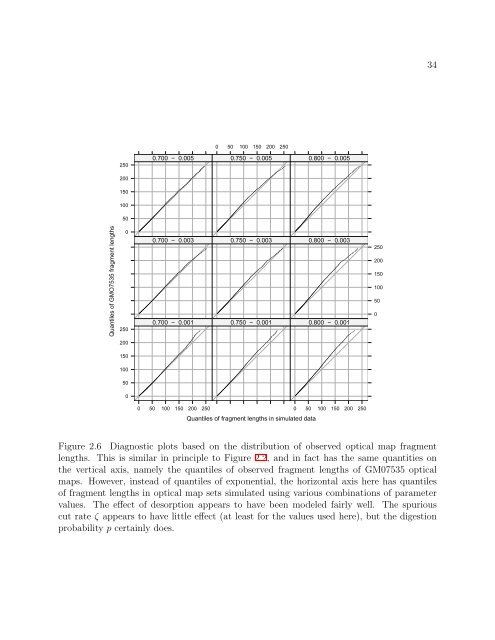
Figure 2.6 Diagnostic plots based on <strong>the</strong> distribution <strong>of</strong> observed optical map fragment<br />
lengths. This is similar in principle to Figure 2.2, and in fact has <strong>the</strong> same quantities on<br />
<strong>the</strong> vertical axis, namely <strong>the</strong> quantiles <strong>of</strong> observed fragment lengths <strong>of</strong> GM07535 optical<br />
maps. However, instead <strong>of</strong> quantiles <strong>of</strong> exponential, <strong>the</strong> horizontal axis here has quantiles<br />
<strong>of</strong> fragment lengths in optical map sets simulated using various combinations <strong>of</strong> parameter<br />
values. The effect <strong>of</strong> desorption appears to have been modeled fairly well. The spurious<br />
cut rate ζ appears to have little effect (at least for <strong>the</strong> values used here), but <strong>the</strong> digestion<br />
probability p certainly does.