Invasive breast carcinoma - IARC
Invasive breast carcinoma - IARC
Invasive breast carcinoma - IARC
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
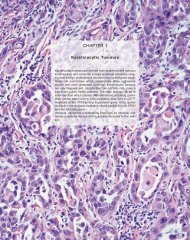
Fig. 1.39 Apocrine <strong>carcinoma</strong>. Note abundant<br />
eosinophilic cytoplasm and vesicular nuclei.<br />
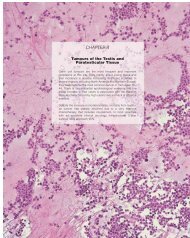
Fig. 1.40 Apocrine <strong>carcinoma</strong>, surperficially resembling<br />
a granular cell tumour.<br />
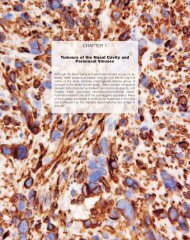
Fig. 1.41 Apocrine <strong>carcinoma</strong>. Immunostaining shows<br />
intense positivity for GCDFP-15.<br />
to as myoblastomatoid {806}. Type B<br />
cell shows abundant cytoplasm in which<br />
fine empty vacuoles are seen. These latter<br />
result in foamy appearance so that<br />
the cells may resemble histiocytes and<br />
sebaceous cells. Nuclei are similar to<br />
those in type A cells. These same cells<br />
have been designated as sebocrine<br />
{2876}. (See also Sebaceous carc i n o-<br />
ma, page 46). C a rcinomas composed<br />
p u rely of foamy apocrine cells may<br />
s u r p e rfically resemble a histiocytic proliferation<br />
or even an inflammatory re a c-<br />
tion {806}. In difficult cases, both granular<br />
cell tumours and histiocytic pro l i f e r a-<br />
tions can be easily distinguished by<br />
staining the tumours with keratin antibodies<br />
that are positive only in apocrine<br />
c a rc i n o m a s .<br />
Immunoprofile<br />
Apocrine <strong>carcinoma</strong>s are typically<br />
GCDFP-15 positive and BCL2 pro t e i n<br />
negative. Expression of GCDFP-15 is a<br />
f e a t u re common to many variants of<br />
b reast <strong>carcinoma</strong>, however, and has<br />
been used to support <strong>breast</strong> origin in<br />
metastatic <strong>carcinoma</strong>s of unknown prim<br />
a ry site. Estrogen and pro g e s t e ro n e<br />
receptors are usually negative in apocrine<br />
<strong>carcinoma</strong> by immonuhistochemical<br />
assessment. Interestingly, many ER-,<br />
PR- apocrine <strong>carcinoma</strong>s do have the<br />
ERmRNA, but fail to produce the pro t e i n<br />
{336}. The expression of other biological<br />
markers is in general similar to that<br />
of other <strong>carcinoma</strong>s {177,1605,2425}.<br />
A n d rogen receptors have been re p o rted<br />
as positive in 97% of ADCIS in one<br />
series {1605} and 81% in another<br />
{2624}. Sixty-two percent of invasive<br />
duct <strong>carcinoma</strong>s were positive in the<br />
latter series {2624} and in 22% of cases<br />
in another study {1874}. The significance<br />
of AR in apocrine <strong>carcinoma</strong>s is<br />
u n c e rt a i n .<br />
G e n e t i c s<br />
Molecular studies in benign, hyperplastic<br />
and neoplastic apocrine lesions parallel<br />
those seen in non apocrine<br />
tumours {1357,1673}.<br />
Prognosis and predictive factors<br />
Survival analysis of 72 cases of invasive<br />
apocrine duct <strong>carcinoma</strong> compared with<br />
non apocrine duct <strong>carcinoma</strong> re v e a l e d<br />
no statistical diff e rence {17,809}.<br />
Metaplastic <strong>carcinoma</strong>s<br />
D e f i n i t i o n<br />
Metaplastic <strong>carcinoma</strong> is a general<br />
t e rm referring to a hetero g e n e o u s<br />
g roup of neoplasms generally characterized<br />
by an intimate admixture of aden<br />
o c a rcinoma with dominant areas of<br />
spindle cell, squamous, and/or mesenchymal<br />
diff e rentiation; the metaplastic<br />
spindle cell and squamous cell <strong>carcinoma</strong>s<br />
may present in a pure form<br />
without any admixture with a re c o g n i z a-<br />
ble adeno<strong>carcinoma</strong>. Metaplastic carc i-<br />
nomas can be classified into bro a d<br />
subtypes according to the phenotypic<br />
appearance of the tumour.<br />
ICD-O code 8 5 7 5 / 3<br />
S y n o n y m s<br />
Matrix producing <strong>carcinoma</strong>, carc i n o-<br />
s a rcoma, spindle cell carc i n o m a .<br />
E p i d e m i o l o g y<br />
Metaplastic <strong>carcinoma</strong>s account for<br />
less than 1% of all invasive mammary<br />
c a rcinomas {1273}. The average age at<br />
p resentation is 55.<br />
Clinical features<br />
Clinical presentation is not diff e rent fro m<br />
that of infiltrating duct NOS carc i n o m a .<br />
Most patients present with a well circ u m-<br />
scribed palpable mass, with a median<br />
size of 3-4 cm, in some re p o rts more<br />
than half of these tumours measure over<br />
5 cm, with some massive lesions (>20<br />
cm) which may displace the nipple and<br />
ulcerate through the skin.<br />
On mammography, most metaplastic<br />
c a rcinomas appear as well delineated<br />
mass densities. Microcalcifications are<br />
not a common feature, but may be pre s-<br />
ent in the adeno<strong>carcinoma</strong>tous are a s ;<br />
ossification, when present, is, of course,<br />
a p p a rent on mammography.<br />
Macroscopy<br />
The tumours are firm, well delineated<br />
and often solid on cut surface. Squamous<br />
or chondroid diff e rentiation is<br />
reflected as pearly white to firm glistening<br />
areas on the cut surface. One large<br />
and/or multiple small cysts may be<br />
a p p a rent on the cut surface of larger<br />
squamous tumours.<br />
Table 1.08<br />
Classification of metaplastic <strong>carcinoma</strong>s.<br />
Purely epithelial<br />
Squamous<br />
Large cell keratinizing<br />
Spindle cell<br />
Acantholytic<br />
Adeno<strong>carcinoma</strong> with spindle cell differentiation<br />
Adenosquamous, including mucoepidermoid<br />
Mixed epithelial and Mesenchymal<br />
(specify components)<br />
Carcinoma with chondroid metaplasia<br />
Carcinoma with osseous metaplasia<br />
Carcinosarcoma (specify components)<br />
<strong>Invasive</strong> <strong>breast</strong> cancer<br />
37