You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
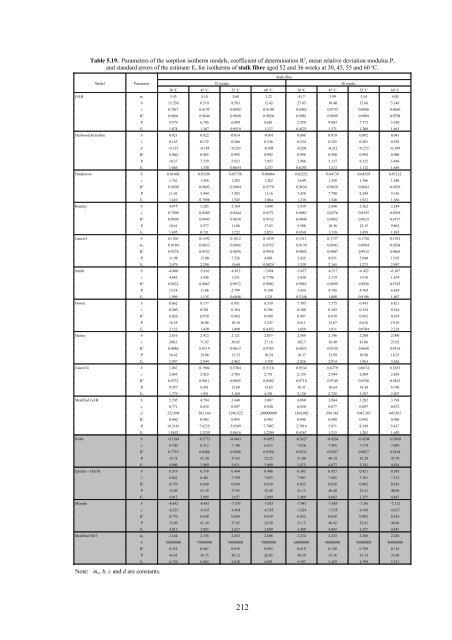
Table 5.19. Parameters of the sorption isotherm models, coefficient of determination R 2 , mean relative deviation modulus P,<br />
and standard errors of the estimate E s for isotherms of stalk fibre aged 52 and 36 weeks at 30, 45, 55 and 60 o C.<br />
Stalk fibre<br />
Model Parameter 52 weeks 36 weeks<br />
30 o C 45 o C 55 o C 60 o C 30 o C 45 o C 55 o C 60 o C<br />
GAB mo 5.43 5.18 3.68 3.22 4.17 3.99 3.54 4.00<br />
b 15.350 6.519 8.583 12.42 27.85 10.40 12.66 5.146<br />
c 0.7817 0.8193 0.8892 0.9140 0.8462 0.8733 0.8886 0.8660<br />
R 2 0.9601 0.9846 0.9949 0.9920 0.9981 0.9899 0.9909 0.9798<br />
P 9.579 6.705 6.099 8.681 2.359 9.083 7.773 5.550<br />
Es 1.874 1.367 0.9519 1.337 0.4523 3.371 1.208 1.663<br />
Hailwood Horrobin b 0.021 0.022 0.014 -0.001 0.006 0.019 0.002 0.041<br />
c 0.143 0.172 0.266 0.336 0.234 0.225 0.301 0.192<br />
d -0.123 -0.158 -0.250 -0.308 -0.204 -0.212 -0.272 -0.199<br />
R 2 0.960 0.985 0.995 0.992 0.998 0.990 0.992 0.980<br />
P 10.37 7.759 5.923 7.857 2.986 5.137 8.152 5.460<br />
Es 1.866 1.330 0.8654 1.257 0.6292 1.213 1.112 1.656<br />
Henderson b 0.01600 0.03501 0.05738 0.06064 0.02252 0.04735 0.04559 0.07212<br />
c 1.761 1.456 1.292 1.262 1.649 1.358 1.386 1.240<br />
R 2 0.9590 0.9893 0.9904 0.9779 0.9830 0.9859 0.9843 0.9939<br />
P 11.01 5.444 7.283 11.16 7.458 7.790 8.294 5.156<br />
Es 1.610 0.7998 1.545 3.004 1.239 1.548 1.923 1.656<br />
Bradley b 4.975 3.285 2.164 1.890 3.539 2.448 2.362 2.244<br />
c 0.7909 0.8209 0.8444 0.8571 0.8085 0.8376 0.8355 0.8309<br />
R 2 0.9590 0.9943 0.9878 0.9732 0.9948 0.9852 0.9815 0.9737<br />
P 10.61 5.577 11.86 17.83 5.588 10.36 12.15 9.862<br />
Es 1.695 0.731 1.252 2.053 0.6541 1.318 1.458 1.693<br />
Caurie I b 0.1401 0.1592 0.1812 0.1839 0.1511 0.1737 0.1720 0.1923<br />
mo 0.9180 0.8833 0.8865 0.8752 0.9139 0.8843 0.8984 0.9204<br />
R 2 0.9378 0.9532 0.9876 0.9958 0.9892 0.9807 0.9915 0.9665<br />
P 11.98 13.90 7.526 4.801 5.425 8.631 5.840 13.05<br />
Es 2.479 2.290 1.644 0.8024 1.339 2.165 1.273 2.997<br />
Smith b -4.900 -5.816 -6.815 -7.494 -5.457 -6.517 -6.422 -6.187<br />
c 4.642 3.436 1.531 0.7796 3.430 2.119 1.918 1.674<br />
R 2 0.9432 0.9863 0.9972 0.9903 0.9965 0.9899 0.9926 0.9745<br />
P 13.14 11.06 2.799 9.100 3.434 4.745 4.769 6.834<br />
Es 1.996 1.135 0.6048 1.231 0.5348 1.088 0.9196 1.667<br />
Oswin b 8.662 8.177 6.951 6.559 7.783 7.375 6.943 6.821<br />
c 0.260 0.301 0.364 0.398 0.300 0.343 0.354 0.344<br />
R 2 0.936 0.978 0.992 0.999 0.987 0.978 0.992 0.955<br />
P 16.35 16.08 10.18 5.247 8.611 13.67 6.616 19.10<br />
Es 2.122 1.430 1.000 0.4352 1.029 1.611 0.9704 2.224<br />
Halsey b 2.816 2.412 2.121 2.057 2.584 2.196 2.260 2.090<br />
c 200.2 71.92 30.65 27.16 102.7 38.40 43.06 23.92<br />
R 2 0.9046 0.9119 0.9613 0.9785 0.9653 0.9534 0.9688 0.9314<br />
P 16.42 19.08 13.25 10.54 10.37 13.99 10.90 18.35<br />
Es 2.997 2.944 2.462 1.758 2.026 2.914 1.963 3.626<br />
Caurie II b 1.067 0.7904 0.5304 0.5116 0.9534 0.6279 0.6674 0.3633<br />
c 2.043 2.423 2.703 2.751 2.154 2.594 2.509 2.824<br />
R 2 0.9733 0.9811 0.9695 0.9492 0.9718 0.9749 0.9556 0.9822<br />
P 9.357 8.451 12.60 15.83 10.31 10.64 14.20 8.790<br />
Es 1.779 1.951 3.169 4.391 2.120 2.725 3.387 2.507<br />
Modified GAB b 5.705 4.784 3.440 3.007 4.080 3.864 3.282 3.798<br />
c 0.771 0.832 0.897 0.920 0.850 0.877 0.897 0.872<br />
d 322.894 503.164 1296.622 200000000 1364.002 694.361 1047.207 443.567<br />
R 2 0.960 0.985 0.995 0.992 0.998 0.990 0.992 0.980<br />
P 10.3181 7.6235 5.9209 7.7407 2.7014 5.071 8.349 5.437<br />
Es 1.8652 1.3230 0.8616 1.2204 0.4367 1.215 1.203 1.650<br />
Kuhn b -0.3184 -0.3773 -0.4441 -0.4953 -0.3627 -0.4264 -0.4204 -0.3849<br />
c 8.700 8.312 7.140 6.813 7.826 7.495 7.174 7.039<br />
R 2 0.7719 0.8408 0.8986 0.9396 0.8533 0.8587 0.9027 0.8194<br />
P 33.75 41.20 37.63 32.25 31.00 40.19 32.20 47.78<br />
Es 4.000 3.869 3.611 3.080 3.475 4.077 3.341 4.434<br />
Iglesias - Chirife b 0.319 0.378 0.444 0.496 0.363 0.427 0.421 0.385<br />
c 8.842 8.481 7.339 7.033 7.987 7.685 7.361 7.212<br />
R 2 0.770 0.840 0.898 0.939 0.852 0.858 0.902 0.818<br />
P 33.89 41.39 37.85 32.49 31.15 40.40 32.41 48.00<br />
Es 4.012 3.885 3.627 3.099 3.489 4.092 3.357 4.447<br />
Mizrahi b -8.842 -8.481 -7.339 -7.033 -7.987 -7.685 -7.361 -7.212<br />
c -8.523 -8.103 -6.894 -6.538 -7.624 -7.258 -6.941 -6.827<br />
R 2 0.770 0.840 0.898 0.939 0.852 0.858 0.902 0.818<br />
P 33.89 41.39 37.85 32.50 31.15 40.42 32.41 48.00<br />
Es 4.012 3.885 3.627 3.099 3.489 4.093 3.357 4.447<br />
Modified BET mo 2.164 2.336 2.453 2.608 2.234 2.433 2.366 2.220<br />
Note: m o, b, c and d are constants.<br />
b 30000000 70000000 50000000 70000000 60000000 40000000 50000000 40000000<br />
R 2 0.354 0.607 0.819 0.893 0.619 0.749 0.799 0.718<br />
P 43.43 36.75 30.12 26.05 39.39 33.36 33.33 33.08<br />
Es 6.730 6.082 4.830 4.091 5.597 5.429 4.799 5.535<br />
212