genomewide characterization of host-pathogen interactions by ...
genomewide characterization of host-pathogen interactions by ...
genomewide characterization of host-pathogen interactions by ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Maren Depke<br />
Results<br />
Host Cell Gene Expression Pattern in an in vitro Infection Model<br />
(1.6), but CASP9, the link to the intrinsic mitochondrial apoptosis pathway, was repressed (−1.6).<br />
Possibly, S9 cells were prepared to react to external apoptosis signals or give signals to infection<br />
relevant immune cells like neutrophils or macrophages. Of course, such situation does not occur<br />
in the S9 cell in vitro infection setting.<br />
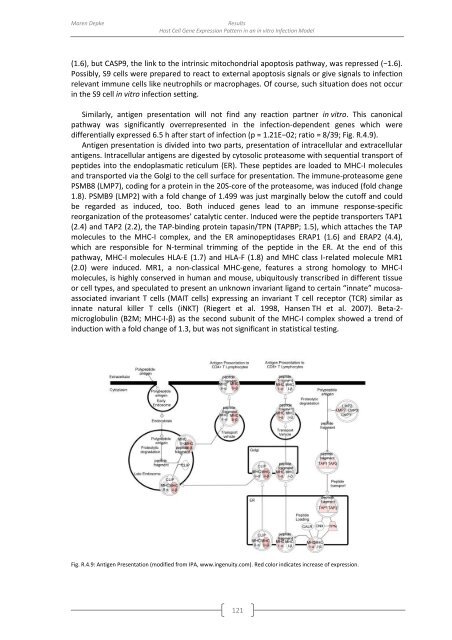
Similarly, antigen presentation will not find any reaction partner in vitro. This canonical<br />
pathway was significantly overrepresented in the infection-dependent genes which were<br />
differentially expressed 6.5 h after start <strong>of</strong> infection (p = 1.21E−02; ratio = 8/39; Fig. R.4.9).<br />
Antigen presentation is divided into two parts, presentation <strong>of</strong> intracellular and extracellular<br />
antigens. Intracellular antigens are digested <strong>by</strong> cytosolic proteasome with sequential transport <strong>of</strong><br />
peptides into the endoplasmatic reticulum (ER). These peptides are loaded to MHC-I molecules<br />
and transported via the Golgi to the cell surface for presentation. The immune-proteasome gene<br />
PSMB8 (LMP7), coding for a protein in the 20S-core <strong>of</strong> the proteasome, was induced (fold change<br />
1.8). PSMB9 (LMP2) with a fold change <strong>of</strong> 1.499 was just marginally below the cut<strong>of</strong>f and could<br />
be regarded as induced, too. Both induced genes lead to an immune response-specific<br />
reorganization <strong>of</strong> the proteasomes’ catalytic center. Induced were the peptide transporters TAP1<br />
(2.4) and TAP2 (2.2), the TAP-binding protein tapasin/TPN (TAPBP; 1.5), which attaches the TAP<br />
molecules to the MHC-I complex, and the ER aminopeptidases ERAP1 (1.6) and ERAP2 (4.4),<br />
which are responsible for N-terminal trimming <strong>of</strong> the peptide in the ER. At the end <strong>of</strong> this<br />
pathway, MHC-I molecules HLA-E (1.7) and HLA-F (1.8) and MHC class I-related molecule MR1<br />
(2.0) were induced. MR1, a non-classical MHC-gene, features a strong homology to MHC-I<br />
molecules, is highly conserved in human and mouse, ubiquitously transcribed in different tissue<br />
or cell types, and speculated to present an unknown invariant ligand to certain “innate” mucosaassociated<br />
invariant T cells (MAIT cells) expressing an invariant T cell receptor (TCR) similar as<br />
innate natural killer T cells (iNKT) (Riegert et al. 1998, Hansen TH et al. 2007). Beta-2-<br />
microglobulin (B2M; MHC-I-β) as the second subunit <strong>of</strong> the MHC-I complex showed a trend <strong>of</strong><br />
induction with a fold change <strong>of</strong> 1.3, but was not significant in statistical testing.<br />
Fig. R.4.9: Antigen Presentation (modified from IPA, www.ingenuity.com). Red color indicates increase <strong>of</strong> expression.<br />
121