genomewide characterization of host-pathogen interactions by ...
genomewide characterization of host-pathogen interactions by ...
genomewide characterization of host-pathogen interactions by ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Maren Depke<br />
Results<br />
Gene Expression Pattern <strong>of</strong> Bone-Marrow Derived Macrophages after Interferon-gamma Treatment<br />
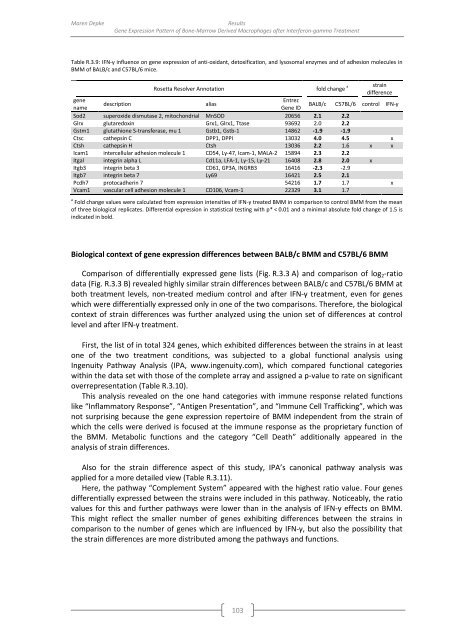
Table R.3.9: IFN-γ influence on gene expression <strong>of</strong> anti-oxidant, detoxification, and lysosomal enzymes and <strong>of</strong> adhesion molecules in<br />
BMM <strong>of</strong> BALB/c and C57BL/6 mice.<br />
Rosetta Resolver Annotation fold change a strain<br />
difference<br />
gene<br />
Entrez<br />
description<br />
alias<br />
name<br />
Gene ID<br />
BALB/c C57BL/6 control IFN-γ<br />
Sod2 superoxide dismutase 2, mitochondrial MnSOD 20656 2.1 2.2<br />
Glrx glutaredoxin Grx1, Glrx1, Ttase 93692 2.0 2.2<br />
Gstm1 glutathione S-transferase, mu 1 Gstb1, Gstb-1 14862 -1.9 -1.9<br />
Ctsc cathepsin C DPP1, DPPI 13032 4.0 4.5 x<br />
Ctsh cathepsin H Ctsh 13036 2.2 1.6 x x<br />
Icam1 intercellular adhesion molecule 1 CD54, Ly-47, Icam-1, MALA-2 15894 2.3 2.2<br />
Itgal integrin alpha L Cd11a, LFA-1, Ly-15, Ly-21 16408 2.8 2.0 x<br />
Itgb3 integrin beta 3 CD61, GP3A, INGRB3 16416 -2.3 -2.9<br />
Itgb7 integrin beta 7 Ly69 16421 2.5 2.1<br />
Pcdh7 protocadherin 7 54216 1.7 1.7 x<br />
Vcam1 vascular cell adhesion molecule 1 CD106, Vcam-1 22329 3.1 1.7<br />
a Fold change values were calculated from expression intensities <strong>of</strong> IFN-γ treated BMM in comparison to control BMM from the mean<br />
<strong>of</strong> three biological replicates. Differential expression in statistical testing with p* < 0.01 and a minimal absolute fold change <strong>of</strong> 1.5 is<br />
indicated in bold.<br />
Biological context <strong>of</strong> gene expression differences between BALB/c BMM and C57BL/6 BMM<br />
Comparison <strong>of</strong> differentially expressed gene lists (Fig. R.3.3 A) and comparison <strong>of</strong> log 2 -ratio<br />
data (Fig. R.3.3 B) revealed highly similar strain differences between BALB/c and C57BL/6 BMM at<br />
both treatment levels, non-treated medium control and after IFN-γ treatment, even for genes<br />
which were differentially expressed only in one <strong>of</strong> the two comparisons. Therefore, the biological<br />
context <strong>of</strong> strain differences was further analyzed using the union set <strong>of</strong> differences at control<br />
level and after IFN-γ treatment.<br />
First, the list <strong>of</strong> in total 324 genes, which exhibited differences between the strains in at least<br />
one <strong>of</strong> the two treatment conditions, was subjected to a global functional analysis using<br />
Ingenuity Pathway Analysis (IPA, www.ingenuity.com), which compared functional categories<br />
within the data set with those <strong>of</strong> the complete array and assigned a p-value to rate on significant<br />
overrepresentation (Table R.3.10).<br />
This analysis revealed on the one hand categories with immune response related functions<br />
like “Inflammatory Response”, “Antigen Presentation”, and “Immune Cell Trafficking”, which was<br />
not surprising because the gene expression repertoire <strong>of</strong> BMM independent from the strain <strong>of</strong><br />
which the cells were derived is focused at the immune response as the proprietary function <strong>of</strong><br />
the BMM. Metabolic functions and the category “Cell Death” additionally appeared in the<br />
analysis <strong>of</strong> strain differences.<br />
Also for the strain difference aspect <strong>of</strong> this study, IPA’s canonical pathway analysis was<br />
applied for a more detailed view (Table R.3.11).<br />
Here, the pathway “Complement System” appeared with the highest ratio value. Four genes<br />
differentially expressed between the strains were included in this pathway. Noticeably, the ratio<br />
values for this and further pathways were lower than in the analysis <strong>of</strong> IFN-γ effects on BMM.<br />
This might reflect the smaller number <strong>of</strong> genes exhibiting differences between the strains in<br />
comparison to the number <strong>of</strong> genes which are influenced <strong>by</strong> IFN-γ, but also the possibility that<br />
the strain differences are more distributed among the pathways and functions.<br />
103