Monitoring mitotic cell division in DT40 cells - Events
Monitoring mitotic cell division in DT40 cells - Events
Monitoring mitotic cell division in DT40 cells - Events
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Tubul<strong>in</strong> DNA<br />
<strong>Monitor<strong>in</strong>g</strong><br />
<strong>mitotic</strong> <strong>cell</strong><br />
<strong>division</strong> <strong>in</strong> <strong>DT40</strong><br />
<strong>cell</strong>s
Our scientific goal: To <strong>in</strong>vestigate the<br />
role of Chk1 <strong>in</strong> chromosome<br />
segregation dur<strong>in</strong>g <strong>mitotic</strong> <strong>cell</strong> <strong>division</strong><br />
• Monitor chromosome segregation dur<strong>in</strong>g <strong>mitotic</strong> <strong>cell</strong> <strong>division</strong><br />
<strong>in</strong> liv<strong>in</strong>g wild-type (wt) and Chk1-/- <strong>DT40</strong> <strong>cell</strong>s by time-lapse<br />
video microscopy<br />
• With time-lapse video microscopy we can monitor events <strong>in</strong><br />
liv<strong>in</strong>g <strong>cell</strong>s as they happen!
Time-lapse video microscopy<br />
• Cells grow <strong>in</strong> appropriate <strong>cell</strong> conta<strong>in</strong>er (e.g. 6-well plate or 30 mm petri dish)<br />
at controlled temperature and CO 2 conditions and are monitored by light<br />
microscopy (phase contrast or DIC)<br />
• If <strong>cell</strong>s express a fluorescence prote<strong>in</strong> of <strong>in</strong>terest they can also be monitored<br />
by fluorescence microscopy<br />
• Cells are monitored for periods up to several hours and a digital photo of<br />
them is taken at desired <strong>in</strong>tervals (e.g. every two m<strong>in</strong>utes, 30 sec, etc)<br />
• If desired, both a fluorescence and a phase contrast/ DIC photo can be taken<br />
at every <strong>in</strong>terval<br />
• At the end of the experiment, part or the whole series of still images can be<br />
compressed <strong>in</strong>to a movie
specimen<br />
Components of a time-lapse video<br />
microscopy system: the microscope<br />
Zeiss, Axionert 200<br />
halogen lamp<br />
mercury<br />
lamp<br />
shutter<br />
objective lenses<br />
• Inverted fluorescence microscope (for<br />
liv<strong>in</strong>g <strong>cell</strong>s)<br />
• Filters for FITC fluorescence<br />
• Objective lenses for DIC or phase<br />
contrast<br />
• Magnification of objective lenses: 4X,<br />
10X, 20X, 40X
Components of a time-lapse video<br />
microscopy system: the temperature<br />
control module<br />
Temperature control module<br />
for 30 mm dishes
Components of a time-lapse video<br />
microscopy system: the environmental<br />
chamber<br />
The environmental<br />
chamber is connected to<br />
a CO 2 source to ma<strong>in</strong>ta<strong>in</strong><br />
appropriate CO 2<br />
conditions (this is<br />
important when <strong>cell</strong>s are<br />
monitored for several<br />
hours)
Components of a time-lapse video<br />
microscopy system: digital camera,<br />
automatic shutter and light beams<br />
• The digital camera is programmed<br />
through a control unit to take photos of the<br />
specimen at predeterm<strong>in</strong>ed <strong>in</strong>tervals for<br />
the period of the experiment<br />
• The microscope shutter automatically<br />
switches from light to fluorescence images<br />
• Both mercury and halogen light beams<br />
automatically switch themselves on before<br />
photos are taken and then switch<br />
themselves off to m<strong>in</strong>imally <strong>in</strong>terfere with<br />
the specimen and avoid photobleach<strong>in</strong>g of<br />
fluorescent molecules<br />
High speed Digital<br />
CCD camera
CO 2 and<br />
temperature<br />
control unit<br />
Overall components of a time-lapse<br />
video microscopy system<br />
Environmental chamber
Generation of <strong>DT40</strong> <strong>cell</strong>s stably<br />
express<strong>in</strong>g H2B:GFP prote<strong>in</strong><br />
• To monitor chromosome segregation, we generated stable clones of wt<br />
and Chk1-/- <strong>DT40</strong> <strong>cell</strong>s express<strong>in</strong>g the chromat<strong>in</strong> marker histone 2B fused<br />
to GFP (H2B:GFP prote<strong>in</strong>)<br />
• Chromosomes <strong>in</strong> those <strong>cell</strong>s can be detected by fluorescence microscopy<br />
(green fluorescence)<br />
General Steps:<br />
1. Obta<strong>in</strong> plasmid cod<strong>in</strong>g for H2B:GFP<br />
2. Electroporate plasmid <strong>in</strong>to <strong>cell</strong>s<br />
3. Select <strong>in</strong>dividual clones express<strong>in</strong>g the plasmid (green clones!)<br />
4. Use clones <strong>in</strong> time-lapse microscopy experiments
Plasmid cod<strong>in</strong>g for H2B:GFP prote<strong>in</strong><br />
Ori<br />
CMV<br />
Promoter<br />
GFP<br />
H2B:GFP<br />
vector<br />
Neomyc<strong>in</strong><br />
Resistance<br />
H2B<br />
Poly-A<br />
Kan R
Electroporation of <strong>DT40</strong> <strong>cell</strong>s (I)<br />
If us<strong>in</strong>g the Amaxa nucleofector system, follow the manufacturer’s<br />
<strong>in</strong>structions!!<br />
Materials and apparatus<br />
• 25 μg of plasmid per 5 x 10 6 <strong>cell</strong>s. (We only l<strong>in</strong>earize plasmid for gene<br />
target<strong>in</strong>g experiments)<br />
• Optimem medium (from Gibco BRL)<br />
• 4 mm electroporation cuvettes (from Flowgen)<br />
• Biorad electroporation micropulser (or similar)
Method<br />
Electroporation of <strong>DT40</strong> <strong>cell</strong>s (II)<br />
• Use 5 x 10 6 <strong>cell</strong>s. Cells must be <strong>in</strong> the log growth phase and I always split them<br />
approx 1:4 the day before electroporation<br />
• On the day of electroporation count <strong>cell</strong>s and sp<strong>in</strong> down 5 x 10 6 <strong>cell</strong>s. Resuspend<br />
<strong>cell</strong>s <strong>in</strong> 1ml of optimem + 1% DMSO. Add plasmid DNA and put the <strong>cell</strong>s + DNA<br />
<strong>in</strong>to an electroporation cuvette<br />
• Electroporate at 300 volts, 960μF<br />
• Dilute <strong>cell</strong>s <strong>in</strong>to 20 ml <strong>DT40</strong> growth medium and <strong>in</strong>cubate O/N at 39.5 o C, 5%<br />
CO 2
Isolation of <strong>DT40</strong> clones<br />
• The next day, take the volume up to 140 ml with <strong>DT40</strong> growth medium<br />
conta<strong>in</strong><strong>in</strong>g the required selection agent (e.g. G418 at 1.5 mg/ ml) and 50 µg/ ml<br />
amphoteric<strong>in</strong> (fungicide)<br />
• Split the 140 ml between 7 x 96 well plates us<strong>in</strong>g a multi-channel pipette (200<br />
μl per well). Incubate at 39.5 o C, 5% CO 2<br />
• After approx 3 weeks exam<strong>in</strong>e the wells for small colonies (balls) of antibiotic<br />
resistant <strong>cell</strong>s. Clones express<strong>in</strong>g H2B:GFP exhibit green fluorescence and are<br />
easily identified!<br />
• Transfer a few clones <strong>in</strong>to 24 well plates and progressively expand them<br />
• Verify DNA fluorescence and that the stages of mitosis look OK by confocal<br />
microscopy. Choose the appropriate clone(s)
Sett<strong>in</strong>g up <strong>DT40</strong>s for time-lapse<br />
microscopy (I)<br />
Prepare coverslips<br />
• Coat 12 mm round coverslips with poly-L-lys<strong>in</strong>e, 10 μg/ ml <strong>in</strong> sterile PBS, O/N,<br />
at 4 o C<br />
• Wash a couple of times with PBS: coverslips are ready to use. You can keep<br />
coated coverslips <strong>in</strong> PBS at 4 o C for a few days<br />
• If <strong>in</strong> a hurry, you can also coat coverslips at 37 o C for 2 hrs and then wash <strong>in</strong><br />
PBS and use
Set up <strong>cell</strong>s<br />
Sett<strong>in</strong>g up <strong>DT40</strong>s for time-lapse<br />
microscopy (II)<br />
• On the previous day, count <strong>cell</strong>s express<strong>in</strong>g H2B:GFP, sp<strong>in</strong> them down and<br />
resuspend <strong>in</strong> fresh medium at approximately 1 x10 5 <strong>cell</strong>s/ ml<br />
• Place a poly-L-lys<strong>in</strong>e-coated coverslip <strong>in</strong>side an empty 30 mm petri dish<br />
• Drop 100 μl (i.e. 1 x 10 4 ) <strong>cell</strong>s onto the coverslip (you don’t want <strong>cell</strong>s too crowded!!)<br />
• Leave at room temperature for approximately 1h for <strong>DT40</strong>s to sit down<br />
• Carefully put 2 ml growth medium <strong>in</strong>side the 30 mm dish and return to the <strong>in</strong>cubator<br />
• Next day, monitor <strong>cell</strong>s on the time-lapse microscope and get still photos or movies!!
<strong>Monitor<strong>in</strong>g</strong> <strong>mitotic</strong> <strong>cell</strong> <strong>division</strong> by time-<br />
lapse microscopy (I)<br />
Photo 1<br />
H2B:GFP DIC<br />
Photo 2<br />
H2B:GFP DIC<br />
Photo 3<br />
H2B:GFP DIC<br />
Photo 4<br />
H2B:GFP DIC
<strong>Monitor<strong>in</strong>g</strong> <strong>mitotic</strong> <strong>cell</strong> <strong>division</strong> by time-<br />
lapse microscopy (II)<br />
H2B:GFP<br />
Photo 5<br />
Photo 6<br />
DIC<br />
H2B:GFP DIC<br />
Photo 7<br />
H2B:GFP DIC<br />
Photo 8<br />
H2B:GFP DIC
<strong>Monitor<strong>in</strong>g</strong> <strong>mitotic</strong> <strong>cell</strong> <strong>division</strong> by time-<br />
lapse video microscopy<br />
DNA DIC
metaphase<br />
anaphase<br />
Normal chromosome<br />
segregation<br />
sp<strong>in</strong>dle pole<br />
sister chromatids<br />
k<strong>in</strong>etochore<br />
metaphase<br />
anaphase<br />
Chromosome missegregation<br />
(polar<br />
chromosome)
Chk1-deficient Chk1 deficient <strong>DT40</strong> <strong>cell</strong>s exhibit high<br />
frequency of “polar polar” chromosomes<br />
wt <strong>DT40</strong> Chk1-/-<br />
Normal anaphase Anaphase with “polar” chromosome<br />
Green: DNA
Chk1-deficient Chk1 deficient <strong>DT40</strong> <strong>cell</strong>s exhibit high<br />
frequency of anaphase bridges<br />
DNA<br />
DNA<br />
Chk1-/-<br />
Chk1-/-<br />
Tubul<strong>in</strong> DNA<br />
Tubul<strong>in</strong> DNA<br />
Chk1-/-<br />
Anaphase bridge<br />
Green: DNA
Chk1-deficient Chk1 deficient <strong>DT40</strong> <strong>cell</strong>s exhibit high<br />
frequency of cytok<strong>in</strong>esis failure<br />
Chk1-/-<br />
Failed cytok<strong>in</strong>esis<br />
Green: DNA
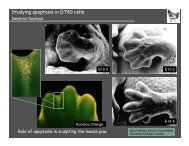
Liv<strong>in</strong>g<br />
<strong>cell</strong><br />
Apoptotic<br />
<strong>cell</strong><br />
Mitotic failure can lead to <strong>cell</strong> death<br />
Electron microscopy<br />
Cell death after <strong>mitotic</strong> failure<br />
Time-lapse microscopy<br />
Green: DNA