Preprint volume - SIBM
Preprint volume - SIBM Preprint volume - SIBM
Pre-print Volume – Posters PLANKTON COMMITEE C. BATTOCCHI, C. TOTTI 1 , M. VILA 2 , M. MASÒ 2 , S. CAPELLACCI, S. ACCORONI 1 , A. RENÉ 2 , M. SCARDI 3 , A. PENNA Dep. of Biomolecular Science, University of Urbino, Viale Trieste, 296 - 61121 Pesaro, Italia. c.battocchi@campus.uniurb.it 1 DISMAR, Università Politecnica delle Marche, Ancona, Italia. 2 ICM-CSIC, Barcelona, Spain. 3 Dep. of Biology, University of Roma “Tor Vergata”, Italia. MONITORING OF TOXIC MICROALGA OSTREOPSIS (DINOFLAGELLATE) SPECIES IN MEDITERRANEAN COASTAL WATERS USING THE PCR BASED-ASSAY COMBINED WITH LIGHT MICROSCOPY MONITORAGGIO DELLA MICROALGA POTENZIALMENTE TOSSICA OSTREOPSIS (DINOFLAGELLATA) IN AREE COSTIERE MEDITERRANEE CON IL METODO MOLECOLARE DI PCR E MICROSCOPIA OTTICA Abstract – A molecular PCR based-assay was developed and applied to macrophyte and seawater samples containing mixed microphytobenthic and phytoplanktonic assemblages in order to detect toxic Ostreopsis species in the Mediterranean Sea. The PCR allowed rapid detection of Ostreopsis cells, even if their abundances are below the light microscopy’s detection limit. Species-specific identification was possible only by PCR-based assay, due to the inherent difficulty of morphological identification in field samples. During the monitoring of the toxic Ostreopsis blooms PCR based methods proved to be effective tools complementary to microscopy for rapid and specific detection of Ostreopsis in marine coastal waters. Key-words: coastal waters, Mediterranean Sea, monitoring, Ostreopsis, PCR. Introduction – Harmful Algal Blooms (HABs) occur frequently in coastal waters throughout the world causing negative impact on environmental quality, human health and economical activities. HAB species include Ostreopsis, a benthic/epiphytic genus known to produce palytoxin-like compounds. Two Ostreopsis species, as O. ovata and O. cf. siamensis, are being found with increasing frequency in the Mediterranean Sea (Battocchi et al., 2010; Totti et al., 2010). Correct identification of these two species is quite difficult by microscopy analyses but it is particular important given that the two species can produce different toxins, which are a potential risk to humans and other organisms. In this study, an efficient PCR based assay was applied to environmental samples in order to monitor the presence of Ostreopsis species in Mediterranean coastal areas, and to compare molecular data with microscopy determinations. Materials and methods – A total of 125 samples of macrophytes, net and surface water samples were collected from April to November 2007 at 20 sites distributed in northern Adriatic Sea and Catalan Sea where blooms of Ostreopsis have never been detected before or commonly occur, respectively. Subsamples were settled for 24h in Sedgwick-Rafter or Utermöhl chambers and Ostreopsis spp. were counted under inverted microscopes (Axiovert 40 CFL and Axiovert 135H, Zeiss, Germany or a Leitz DM-II, Germany) at 200× or 400× magnification on the half or entire sedimentation chamber. Pellets obtained from macrophyte and seawater samples were used for total DNA extraction and purification according to Battocchi et al. (2010). Genus and species-specific primers were designed in the 5.8S rDNA-ITS regions. PCR reactions were carried out directly using these primers or by following two steps: an initial PCR 41 st S.I.B.M. CONGRESS Rapallo (GE), 7-11 June 2010 333
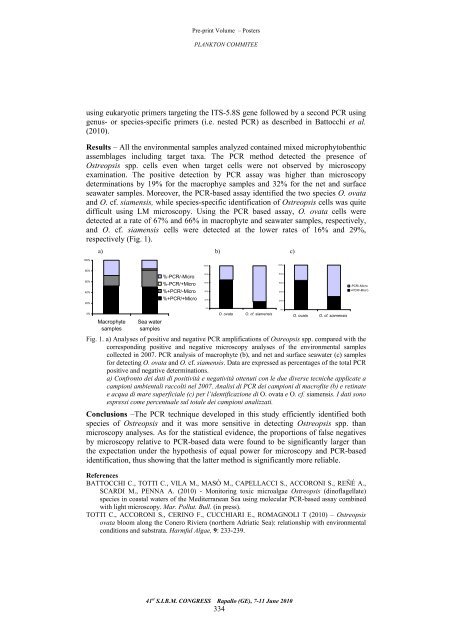
Pre-print Volume – Posters PLANKTON COMMITEE using eukaryotic primers targeting the ITS-5.8S gene followed by a second PCR using genus- or species-specific primers (i.e. nested PCR) as described in Battocchi et al. (2010). Results – All the environmental samples analyzed contained mixed microphytobenthic assemblages including target taxa. The PCR method detected the presence of Ostreopsis spp. cells even when target cells were not observed by microscopy examination. The positive detection by PCR assay was higher than microscopy determinations by 19% for the macrophye samples and 32% for the net and surface seawater samples. Moreover, the PCR-based assay identified the two species O. ovata and O. cf. siamensis, while species-specific identification of Ostreopsis cells was quite difficult using LM microscopy. Using the PCR based assay, O. ovata cells were detected at a rate of 67% and 66% in macrophyte and seawater samples, respectively, and O. cf. siamensis cells were detected at the lower rates of 16% and 29%, respectively (Fig. 1). 100% 80% 60% 40% 20% 0% a) b) c) Macrophyte samples Sea water samples %-PCR/-Micro %-PCR/+Micro %+PCR/-Micro %+PCR/+Micro 100% 80% 60% 40% 20% 0% O. ovata O. cf. siamensis Fig. 1. a) Analyses of positive and negative PCR amplifications of Ostreopsis spp. compared with the corresponding positive and negative microscopy analyses of the environmental samples collected in 2007. PCR analysis of macrophyte (b), and net and surface seawater (c) samples for detecting O. ovata and O. cf. siamensis. Data are expressed as percentages of the total PCR positive and negative determinations. a) Confronto dei dati di positività e negatività ottenuti con le due diverse tecniche applicate a campioni ambientali raccolti nel 2007. Analisi di PCR dei campioni di macrofite (b) e retinate e acqua di mare superficiale (c) per l’identificazione di O. ovata e O. cf. siamensis. I dati sono espressi come percentuale sul totale dei campioni analizzati. Conclusions –The PCR technique developed in this study efficiently identified both species of Ostreopsis and it was more sensitive in detecting Ostreopsis spp. than microscopy analyses. As for the statistical evidence, the proportions of false negatives by microscopy relative to PCR-based data were found to be significantly larger than the expectation under the hypothesis of equal power for microscopy and PCR-based identification, thus showing that the latter method is significantly more reliable. References BATTOCCHI C., TOTTI C., VILA M., MASÓ M., CAPELLACCI S., ACCORONI S., REÑÉ A., SCARDI M., PENNA A. (2010) - Monitoring toxic microalgae Ostreopsis (dinoflagellate) species in coastal waters of the Mediterranean Sea using molecular PCR-based assay combined with light microscopy. Mar. Pollut. Bull. (in press). TOTTI C., ACCORONI S., CERINO F., CUCCHIARI E., ROMAGNOLI T (2010) – Ostreopsis ovata bloom along the Conero Riviera (northern Adriatic Sea): relationship with environmental conditions and substrata. Harmful Algae, 9: 233-239. 41 st S.I.B.M. CONGRESS Rapallo (GE), 7-11 June 2010 334 100% 80% 60% 40% 20% 0% O. ovata O. cf. siamensis -PCR/-Micro +PCR/-Micro
- Page 284 and 285: Pre-print Volume - Posters BENTHOS
- Page 286 and 287: Pre-print Volume - Posters BENTHOS
- Page 288 and 289: Pre-print Volume - Posters BENTHOS
- Page 290 and 291: Pre-print Volume - Posters MANAGEME
- Page 292 and 293: Pre-print Volume - Posters MANAGEME
- Page 294 and 295: Pre-print Volume - Posters MANAGEME
- Page 296 and 297: Pre-print Volume - Posters MANAGEME
- Page 298 and 299: Pre-print Volume - Posters MANAGEME
- Page 300 and 301: Pre-print Volume - Posters MANAGEME
- Page 302 and 303: Pre-print Volume - Posters MANAGEME
- Page 304 and 305: Pre-print Volume - Posters MANAGEME
- Page 306 and 307: Pre-print Volume NECTON AND FISHERY
- Page 308 and 309: Pre-print Volume -Posters NECTON AN
- Page 310 and 311: Pre-print Volume -Posters NECTON AN
- Page 312 and 313: Pre-print Volume -Posters NECTON AN
- Page 314 and 315: Pre-print Volume -Posters NECTON AN
- Page 316 and 317: Pre-print Volume -Posters NECTON AN
- Page 318 and 319: Pre-print Volume -Posters NECTON AN
- Page 320 and 321: Pre-print Volume -Posters NECTON AN
- Page 322 and 323: Pre-print Volume -Posters NECTON AN
- Page 324 and 325: Pre-print Volume -Posters NECTON AN
- Page 326 and 327: Pre-print Volume -Posters NECTON AN
- Page 328 and 329: Pre-print Volume -Posters NECTON AN
- Page 330 and 331: Pre-print Volume -Posters NECTON AN
- Page 332 and 333: Pre-print Volume -Posters NECTON AN
- Page 336 and 337: Pre-print Volume - Posters PLANKTON
- Page 338 and 339: Pre-print Volume - Posters PLANKTON
- Page 340 and 341: Pre-print Volume - Posters PLANKTON
- Page 342 and 343: Pre-print Volume - Posters PLANKTON
- Page 344 and 345: Pre-print Volume - Posters PLANKTON
- Page 346 and 347: Pre-print Volume VARIOUS TOPICS VAR
- Page 348 and 349: Pre-print Volume - Posters VARIOUS
- Page 350 and 351: Pre-print Volume - Posters VARIOUS
- Page 352 and 353: Pre-print Volume - Posters VARIOUS
- Page 354 and 355: Pre-print Volume - Posters VARIOUS
- Page 356 and 357: Pre-print Volume - Posters VARIOUS
- Page 358 and 359: Pre-print Volume - Posters VARIOUS
- Page 360 and 361: Pre-print Volume - Posters VARIOUS
- Page 362 and 363: Pre-print Volume - Posters VARIOUS
- Page 364 and 365: Pre-print Volume - Posters VARIOUS
- Page 366 and 367: Pre-print Volume - Posters VARIOUS
- Page 368 and 369: Pre-print Volume - Posters VARIOUS
- Page 370 and 371: Castellano M.......................
- Page 372 and 373: Maravelias C.......................
- Page 374: Traversi I. .......................
Pre-print Volume – Posters<br />
PLANKTON COMMITEE<br />
using eukaryotic primers targeting the ITS-5.8S gene followed by a second PCR using<br />
genus- or species-specific primers (i.e. nested PCR) as described in Battocchi et al.<br />
(2010).<br />
Results – All the environmental samples analyzed contained mixed microphytobenthic<br />
assemblages including target taxa. The PCR method detected the presence of<br />
Ostreopsis spp. cells even when target cells were not observed by microscopy<br />
examination. The positive detection by PCR assay was higher than microscopy<br />
determinations by 19% for the macrophye samples and 32% for the net and surface<br />
seawater samples. Moreover, the PCR-based assay identified the two species O. ovata<br />
and O. cf. siamensis, while species-specific identification of Ostreopsis cells was quite<br />
difficult using LM microscopy. Using the PCR based assay, O. ovata cells were<br />
detected at a rate of 67% and 66% in macrophyte and seawater samples, respectively,<br />
and O. cf. siamensis cells were detected at the lower rates of 16% and 29%,<br />
respectively (Fig. 1).<br />
100%<br />
80%<br />
60%<br />
40%<br />
20%<br />
0%<br />
a) b) c)<br />
Macrophyte<br />
samples<br />
Sea water<br />
samples<br />
%-PCR/-Micro<br />
%-PCR/+Micro<br />
%+PCR/-Micro<br />
%+PCR/+Micro<br />
100%<br />
80%<br />
60%<br />
40%<br />
20%<br />
0%<br />
O. ovata O. cf. siamensis<br />
Fig. 1. a) Analyses of positive and negative PCR amplifications of Ostreopsis spp. compared with the<br />
corresponding positive and negative microscopy analyses of the environmental samples<br />
collected in 2007. PCR analysis of macrophyte (b), and net and surface seawater (c) samples<br />
for detecting O. ovata and O. cf. siamensis. Data are expressed as percentages of the total PCR<br />
positive and negative determinations.<br />
a) Confronto dei dati di positività e negatività ottenuti con le due diverse tecniche applicate a<br />
campioni ambientali raccolti nel 2007. Analisi di PCR dei campioni di macrofite (b) e retinate<br />
e acqua di mare superficiale (c) per l’identificazione di O. ovata e O. cf. siamensis. I dati sono<br />
espressi come percentuale sul totale dei campioni analizzati.<br />
Conclusions –The PCR technique developed in this study efficiently identified both<br />
species of Ostreopsis and it was more sensitive in detecting Ostreopsis spp. than<br />
microscopy analyses. As for the statistical evidence, the proportions of false negatives<br />
by microscopy relative to PCR-based data were found to be significantly larger than<br />
the expectation under the hypothesis of equal power for microscopy and PCR-based<br />
identification, thus showing that the latter method is significantly more reliable.<br />
References<br />
BATTOCCHI C., TOTTI C., VILA M., MASÓ M., CAPELLACCI S., ACCORONI S., REÑÉ A.,<br />
SCARDI M., PENNA A. (2010) - Monitoring toxic microalgae Ostreopsis (dinoflagellate)<br />
species in coastal waters of the Mediterranean Sea using molecular PCR-based assay combined<br />
with light microscopy. Mar. Pollut. Bull. (in press).<br />
TOTTI C., ACCORONI S., CERINO F., CUCCHIARI E., ROMAGNOLI T (2010) – Ostreopsis<br />
ovata bloom along the Conero Riviera (northern Adriatic Sea): relationship with environmental<br />
conditions and substrata. Harmful Algae, 9: 233-239.<br />
41 st S.I.B.M. CONGRESS Rapallo (GE), 7-11 June 2010<br />
334<br />
100%<br />
80%<br />
60%<br />
40%<br />
20%<br />
0%<br />
O. ovata O. cf. siamensis<br />
-PCR/-Micro<br />
+PCR/-Micro