Molecular detection of poisonous mushrooms in ... - Mycologia
Molecular detection of poisonous mushrooms in ... - Mycologia
Molecular detection of poisonous mushrooms in ... - Mycologia
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>Mycologia</strong>, 102(3), 2010, pp. 747–754. DOI: 10.3852/09-124<br />
# 2010 by The Mycological Society <strong>of</strong> America, Lawrence, KS 66044-8897<br />
Sara Epis 1<br />
<strong>Molecular</strong> <strong>detection</strong> <strong>of</strong> <strong>poisonous</strong> <strong>mushrooms</strong> <strong>in</strong> different matrices<br />
Dipartimento di Patologia Animale, Igiene e Sanità<br />
Pubblica Veter<strong>in</strong>aria, Università degli studi di Milano,<br />
Milano, Italy<br />
Cater<strong>in</strong>a Mat<strong>in</strong>ato<br />
Responsabile U.O. Preparazione terreni di coltura<br />
Laboratorio di Sanità Pubblica, ASL di Milano, Italy<br />
Gabriella Gentili<br />
Responsabile U.O. Microbiologia Cl<strong>in</strong>ica, Sezione<br />
Specialistica di Micologia, Laboratorio di Sanità<br />
Pubblica, ASL di Milano, Italy<br />
Fabio Varotto<br />
Responsabile Servizio medico, Laboratorio di Sanità<br />
Pubblica, ASL di Milano, Italy<br />
Claudio Bandi<br />
Davide Sassera2 Dipartimento di Patologia Animale, Igiene e Sanità<br />
Pubblica Veter<strong>in</strong>aria, Università degli studi di Milano,<br />
Milano, Italy<br />
Abstract: Amanita phalloides, Lepiota cristata, Lepiota<br />
brunneo<strong>in</strong>carnata and Inocybe asterospora are among<br />
the most important species responsible <strong>of</strong> mushroom<br />
poison<strong>in</strong>g <strong>in</strong> northern Italy. A real time PCR method<br />
for the identification <strong>of</strong> samples conta<strong>in</strong><strong>in</strong>g DNA<br />
from each <strong>of</strong> these species was developed. To test<br />
specificity all protocols were applied on DNA extracted<br />
from various mushroom species; sensitivity was<br />
assessed perform<strong>in</strong>g serial dilutions on all samples;<br />
versatility <strong>of</strong> the protocols was evaluated perform<strong>in</strong>g<br />
tests on DNA extracted from different matrices. The<br />
protocols showed high sensitivity (32 ng dried<br />
mushroom), high specificity and sensitive <strong>detection</strong><br />
<strong>of</strong> DNA extracted from difficult samples, <strong>in</strong>clud<strong>in</strong>g<br />
pasta with mushroom, cooked <strong>mushrooms</strong> and gastric<br />
aspirates.<br />
Key words: Basidiomycetes, <strong>poisonous</strong> <strong>mushrooms</strong>,<br />
real time PCR, species identification<br />
INTRODUCTION<br />
Poisonous <strong>mushrooms</strong> are rout<strong>in</strong>ely identified by<br />
specialized mycologists based on their morphological<br />
characters, but the samples exam<strong>in</strong>ed <strong>in</strong> the context<br />
Submitted 14 Sep 2009; accepted for publication 27 Oct 2009.<br />
1 Current address: Dipartimento di Medic<strong>in</strong>a Sperimentale e Sanità<br />
Pubblica, Università degli Studi di Camer<strong>in</strong>o, Camer<strong>in</strong>o, Italy.<br />
2 Correspond<strong>in</strong>g author. E-mail: davide.sassera@unimi.it<br />
747<br />
<strong>of</strong> cl<strong>in</strong>ical diagnosis are generally not well preserved<br />
and thus frequently unsuitable for a rapid morphological<br />
identification. Furthermore morphological<br />
exam<strong>in</strong>ation is time consum<strong>in</strong>g and requires the<br />
pr<strong>of</strong>essional knowledge <strong>of</strong> a mycologist. The analysis<br />
<strong>of</strong> cooked <strong>mushrooms</strong> or <strong>of</strong> gastric aspirates from<br />
poisoned patients is particularly difficult because the<br />
spores are <strong>of</strong>ten few and their morphology generally<br />
is altered (Hall et al. 1987, Barbato 1993, McPartland<br />
et al. 1997). This paper presents novel primer pairs<br />
and a real time PCR method for the <strong>detection</strong> <strong>of</strong> four<br />
species <strong>of</strong> <strong>poisonous</strong> <strong>mushrooms</strong> that are a common<br />
cause <strong>of</strong> human <strong>in</strong>toxication <strong>in</strong> Italy, Amanita<br />
phalloides, Lepiota cristata, Lepiota brunneo<strong>in</strong>carnata<br />
and Inocybe asterospora.<br />
In Italy 1996–2006 about 10 000 cases <strong>of</strong> mushroom<br />
poison<strong>in</strong>g were reported to the Centro Antiveleni <strong>of</strong><br />
Milano. About 2400 <strong>of</strong> these cases showed a long<br />
<strong>in</strong>cubation period; 22 cases resulted <strong>in</strong> the death <strong>of</strong><br />
the patients; and n<strong>in</strong>e required liver transplants due<br />
to severe hepatic <strong>in</strong>sufficiency (Assisi et al. 2008). In<br />
northern Italy 2005–2008 15% cases have been caused<br />
by mushroom genera Lepiota, Amanita and Inocybe<br />
(data recorded at the Mycology Section, Laboratorio<br />
di Sanità Pubblica <strong>of</strong> Milano, ASL Via F. Juvara, 22–<br />
20129 Milano).<br />
Toxic <strong>mushrooms</strong> <strong>of</strong> genus Lepiota <strong>of</strong>ten are<br />
mistaken for the edible <strong>mushrooms</strong> <strong>of</strong> genus Macrolepiota<br />
and thus are common cause <strong>of</strong> poison<strong>in</strong>g.<br />
Similarly A. phalloides can be misidentified as an<br />
edible species <strong>of</strong> genera Amanita, Lepiota or Russula,<br />
thus caus<strong>in</strong>g 8% <strong>of</strong> the total fungal poison<strong>in</strong>gs <strong>in</strong> Italy<br />
(Assisi et al. 2008). I. asterospora is widespread <strong>in</strong><br />
northern Italy (data recorded by the Associazione<br />
Micologica Bresadola, Varese; http://digilander.libero.<br />
it/ambvarese/<strong>in</strong>dex.htm) and can be misidentified as<br />
Armillaria spp. The above <strong>poisonous</strong> fungi are characterized<br />
by different toxicity; A. phalloides, L. cristata and<br />
L. brunneo<strong>in</strong>carnata cause severe poison<strong>in</strong>g characterized<br />
by a lengthy <strong>in</strong>cubation (Karlson-Stiber et al. 2003,<br />
Roux et al. 2008); I. asterospora causes milder symptoms<br />
with short latency, from 15 m<strong>in</strong> to 4 h (Lurie et al.<br />
2009).<br />
The availability <strong>of</strong> molecular techniques for the<br />
identification <strong>of</strong> <strong>poisonous</strong> fungi would support and<br />
<strong>in</strong>tegrate the work <strong>of</strong> the mycologist <strong>in</strong> cases <strong>of</strong><br />
cl<strong>in</strong>ical poison<strong>in</strong>g. In this study we developed a rapid<br />
system for identification <strong>of</strong> four species <strong>of</strong> <strong>poisonous</strong><br />
mushroom with real time PCR. We show the<br />
application <strong>of</strong> this technique on cooked <strong>mushrooms</strong><br />
and gastric aspirates. The described technique allows
748 MYCOLOGIA<br />
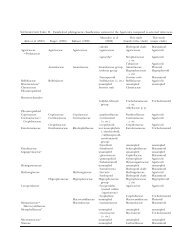
TABLE I. Samples used <strong>in</strong> this study, accession number <strong>of</strong> ITS gene sequences from each species, place and year <strong>of</strong> collection <strong>of</strong> the samples and identification<br />
Year <strong>of</strong><br />
isolation Identification<br />
ITS gene bank<br />
accession number Source Geography Collector<br />
Isolate species<br />
Amanita phalloides EU909444 LSP-MI-68 Parco delle Groane, Milano,<br />
G. Gentili 2007 molecular & standard<br />
Lombardia, Italy<br />
taxonomic keys<br />
Amanita phalloides EU909444 LSP-MI-CS17 Livigno, Sondrio, Lombardia, Italy Gr. Micologico Agrate 2008 standard taxonomic keys<br />
Brianza<br />
Amanita phalloides EU909444 LSP-MI-CS21 Clusone, Bergamo, Lombardia, Italy Gr. Micologico Agrate 2007 standard taxonomic keys<br />
Brianza<br />
Amanita virosa FJ755188 LSP-MI-79 Bellamonte, Trento, Trent<strong>in</strong>o, Italy G. Gentili 1993 molecular & standard<br />
taxonomic keys<br />
Amanita virosa FJ755188 LSP-MI-CS1 Valbondione, Bergamo, Lombardia, Gr. Micologico Agrate 2008 standard taxonomic keys<br />
Italy<br />
Brianza<br />
Amanita verna EU909448 LSP-MI-78 Dossena, Bergamo, Lombardia, Italy G. Gentili 1994 molecular & standard<br />
taxonomic keys<br />
Amanita verna EU909448 LSP-MI-CS11 San Rossore, Pisa, Lombardia, Italy Gr. Micologico Agrate 2006 standard taxonomic keys<br />
Brianza<br />
Amanita cesarea AY486237 LSP-MI-50 Parco delle Groane, Milano,<br />
G. Gentili 2003 standard taxonomic keys<br />
Lombardia, Italy<br />
Amanita muscaria GQ267469 LSP-MI-CS10 Barni, Como, Lombardia, Italy Gr. Micologico Agrate 2000 standard taxonomic keys<br />
Brianza<br />
Amanita panther<strong>in</strong>a GQ401354 LSP-MI-CS15 Triuggio, Monza e Brianza,<br />
Gr. Micologico Agrate 2000 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Lepiota lilacea AY176379 LSP-MI-768 Parco delle Groane, Milano,<br />
G. Gentili 2000 standard taxonomic keys<br />
Lombardia, Italy<br />
Lepiota cristata AJ237628 LSP-MI-757 Rozzano, Milano, Lombardia, Italy G. Gentili 1996 molecular & standard<br />
taxonomic keys<br />
Lepiota cristata AJ237628 LSP-MI-CS22 Cologno Monzese, Milano,<br />
Gr. Micologico Agrate 2006 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Lepiota<br />
FJ481017 LSP-MI-750 Rozzano, Milano, Lombardia, Italy G. Gentili 1996 molecular & standard<br />
brunneo<strong>in</strong>carnata<br />
taxonomic keys<br />
Lepiota<br />
FJ481017 LSP-MI-CS2 Cologno Monzese, Milano,<br />
Gr. Micologico Agrate 2005 standard taxonomic keys<br />
brunneo<strong>in</strong>carnata<br />
Lombardia, Italy<br />
Brianza<br />
Lepiota sub<strong>in</strong>carnata AY176491 LSP-MI-CS3 Cologno Monzese, Milano,<br />
Gr. Micologico Agrate 2004 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Lepiota josserandii ND LSP-MI-765 Parco delle Groane, Milano,<br />
G. Gentili 1996 standard taxonomic keys<br />
Lombardia, Italy
TABLE I. Cont<strong>in</strong>ued.<br />
EPIS ET AL.: POISONOUS MUSHROOMS 749<br />
ITS gene bank<br />
Year <strong>of</strong><br />
Isolate species accession number Source Geography Collector isolation Identification<br />
Russula heterophylla DQ422006 LSP-MI-CS6 Arcore, Monza e Brianza,<br />
Gr. Micologico Agrate 2007 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Marasmius oreades FJ431267 LSP-MI-CS7 Vimercate, Monza e Brianza,<br />
Gr. Micologico Agrate 2005 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Leucoagaricus<br />
AF482865 LSP-MI-CS8 Rozzano, Milano, Lombardia, Italy Gr. Micologico Agrate 2005 standard taxonomic keys<br />
leucothites<br />
Brianza<br />
Inocybe asterospora AM882897 LSP-MI-613 Trent<strong>in</strong>o, Italy G. Gentili 1996 molecular & standard<br />
taxonomic keys<br />
Inocybe asterospora AM882897 LSP-MI-CS24 Barni, Como, Lombardia, Italy Gr. Micologico Agrate 2002 standard taxonomic keys<br />
Brianza<br />
Agaricus xanthodermus DQ182534 LSP-MI-36 Parco delle Groane, Milano,<br />
G. Gentili 1994 molecular & standard<br />
Lombardia, Italy<br />
taxonomic keys<br />
Agaricus arvensis AF161015 LSP-MI-CS9 Cass<strong>in</strong>a de Pecchi, Milano,<br />
Gr. Micologico Agrate 2005 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Agaricus campestris FJ755230 LSP-MI-CS14 Cass<strong>in</strong>a de Pecchi, Milano,<br />
Gr. Micologico Agrate 2006 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Cort<strong>in</strong>arius orellanus AF389164 LSP-MI-CS5 Clusone, Bergamo, Lombardia, Italy Gr. Micologico Agrate 2005 standard taxonomic keys<br />
Brianza<br />
Cort<strong>in</strong>arius<br />
AF261501 LSP-MI-CS12 Aprica, Sondrio, Lombardia, Italy Gr. Micologico Agrate 2005 standard taxonomic keys<br />
speciosissimus<br />
Brianza<br />
Cort<strong>in</strong>arius<br />
AF261501 LSP-MI-CS25 Bossico, Bergamo, Lombardia, Italy Gr. Micologico Agrate 2006 standard taxonomic keys<br />
speciosissimus<br />
Brianza<br />
Chroogomphus AF205650 LSP-MI-CS13 Livigno, Sondrio, Lombardia, Italy Gr. Micologico Agrate 2006 standard taxonomic keys<br />
helveticus<br />
Brianza<br />
Chroogomphus rutilus DQ367894 LSP-MI-CS19 Vimercate, Monza e Brianza,<br />
Gr. Micologico Agrate 2006 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Tricholoma equestre EF493263 LSP-MI-CS16 Chiesa Val Malenco, Sondrio, Gr. Micologico Agrate 2007 standard taxonomic keys<br />
Lombardia, Italy<br />
Brianza<br />
Entoloma lividum AF261294 LSP-MI-CS20 Milano, Lombardia, Italy Gr. Micologico Agrate 2000 standard taxonomic keys<br />
Brianza<br />
Armillaria mellea FJ664597 LSP-MI-82 Clusone, Bergamo, Lombardia, Italy G. Gentili 2000 standard taxonomic keys<br />
Armillaria mellea FJ664597 LSP-MI-CS23 Clusone, Bergamo, Lombardia, Italy Gr. Micologico Agrate 2006 standard taxonomic keys<br />
Brianza<br />
Armillaria ostoyae EU257717 LSP-MI-CS4 Clusone, Bergamo, Lombardia, Italy Gr. Micologico Agrate 2008 standard taxonomic keys<br />
Brianza
750 MYCOLOGIA<br />
TABLE I. Cont<strong>in</strong>ued.<br />
Year <strong>of</strong><br />
isolation Identification<br />
ITS gene bank<br />
accession number Source Geography Collector<br />
Isolate species<br />
Macrolepiota procera AM946456 LSP-MI-772 Parco delle Groane, Milano,<br />
G. Gentili 2006 standard taxonomic keys<br />
Lombardia, Italy<br />
Galer<strong>in</strong>a marg<strong>in</strong>ata AY228347 LSP-MI-457 Perg<strong>in</strong>e, Trento, Trent<strong>in</strong>o, Italy G. Gentili 1994 molecular & standard<br />
taxonomic keys<br />
Boletus calopus DQ679806 LSP-MI-106 Valtell<strong>in</strong>a, Sondrio, Lombardia, Italy G. Gentili 1996 standard taxonomic keys<br />
Gyromitra esculenta AJ544209 LSP-MI-CS18 Sommalombardo, Varese, Lombardia, Gr. Micologico Agrate 2006 standard taxonomic keys<br />
Italy<br />
Brianza<br />
Morchella esculenta EU600241 LSP-MI-750 Ballabio, Lecco, Lombardia, Italy G. Gentili 2006 standard taxonomic keys<br />
ND: Not done.<br />
LSP-MI & LSP-MI-CS: Collection deposited <strong>in</strong> the herbarium <strong>of</strong> the ‘‘Laboratorio di Sanità Pubblica’’, <strong>of</strong> Milano, Italy.<br />
for rapid, sensitive and specific identification <strong>of</strong> four<br />
mushroom species that frequently are recorded as<br />
be<strong>in</strong>g responsible for poison<strong>in</strong>gs <strong>in</strong> northern Italy.<br />
MATERIALS AND METHODS<br />
DNA extraction.—Forty mushroom samples from 31 species<br />
<strong>of</strong> Basidiomycetes have been collected <strong>in</strong> northern Italy<br />
1993–2008 from 23 locations (TABLE I). All samples were<br />
identified morphologically with standard taxonomic keys<br />
(Moser 1993, Mazza 1995, Bon 1988) then subjected to<br />
rapid dry<strong>in</strong>g (<strong>in</strong> a food dehydrator). Ten milligrams <strong>of</strong> each<br />
sample were used for DNA extraction. Samples were put <strong>in</strong><br />
cryotubes and left 10 m<strong>in</strong> <strong>in</strong> liquid nitrogen, then disrupted<br />
by rapid vibration <strong>in</strong> Ribolyser (Hybaid, Middlesex, UK)<br />
with two sizes <strong>of</strong> microbeads (1 mm and 100 mm polystyrene<br />
beads). DNA was extracted with DNeasy Plant M<strong>in</strong>i Kit<br />
(QIAGEN, Hilden, Germany) accord<strong>in</strong>g to manufacturer<br />
<strong>in</strong>structions. DNA was stored at 220 C for molecular<br />
analyses. The above protocol for DNA extraction also was<br />
used on 30 samples <strong>of</strong> gastric aspirates and on 10 samples <strong>of</strong><br />
cooked <strong>mushrooms</strong> collected by the Laboratorio di Sanità<br />
Pubblica <strong>of</strong> Milano. Morphological identification on these<br />
cl<strong>in</strong>ical and cooked samples was possible only <strong>in</strong> a few cases.<br />
(See TABLE I for a description all exam<strong>in</strong>ed samples.)<br />
Primer design and PCR.—DNA quality was tested for each<br />
sample via spectrophotometer (NanoDrop ND 1000,<br />
Thermo Fisher Scientific, Wilm<strong>in</strong>gton, Delaware) and with<br />
two PCR protocols that amplify respectively the <strong>in</strong>ternal<br />
transcribed spacer 1 and 2 (ITS1-ITS2) fragments from all<br />
Basidiomycetes species (Withe et al. 1990, Palapala et al.<br />
2002). The ITS regions are highly conserved with<strong>in</strong> most<br />
species but are variable among species and thus are useful<br />
<strong>in</strong> taxonomy and for sensitive and selective species<br />
identification (Turenne et al. 2000).<br />
Four primer pairs (TABLE II) were manually designed<br />
based on an alignment <strong>of</strong> ITS1-5S rDNA-ITS2 gene sequences<br />
from 40 species <strong>of</strong> Basidiomycetes (Bao et al. 2005, Schmidt et<br />
al. 2000), generated with Clustal X 2.0 (Lark<strong>in</strong> et al. 2007).<br />
These primer pairs were designed on ITS1 or ITS2 (see<br />
TABLE II) to be species specific for each <strong>of</strong> the follow<strong>in</strong>g: A.<br />
phalloides, L. cristata, L. brunneo<strong>in</strong>carnata and I. asterospora.<br />
Primer specificity then was checked by BLAST analyses.<br />
Each primer pair was tested for PCR specificity with DNA<br />
from each <strong>of</strong> the 40 mushroom species with the same<br />
amplification conditions: 18 mL buffer (10 mM Tris-HCl<br />
[pH 8.3], 50 mM KCl, 1.5 mM MgCl 2) with 0.2 mM each<br />
deoxynucleoside triphosphate, 1 mM each primer, 0.5 U Taq<br />
Polymerase (Euroclone) and 2 mL DNA extraction obta<strong>in</strong>ed<br />
as above. The thermal pr<strong>of</strong>ile was 2 m<strong>in</strong> at 90 C; 35 cycles <strong>of</strong><br />
95 C for 40 s, 57 C for 40 s and 72 C for 40 s; the elongation<br />
was completed with 10 m<strong>in</strong> at 72 C.<br />
PCR products were gel-purified with the QIAquick TM Gel<br />
Extraction Kit (QIAGEN) accord<strong>in</strong>g to manufacturer<br />
protocols, resuspended <strong>in</strong> 30 mL deionized water and<br />
sequenced with PCR primers with the ABI PRISM BigDye<br />
Term<strong>in</strong>ator Cycle Sequenc<strong>in</strong>g Reaction Kit 3.1 (Applera<br />
Europe, Warr<strong>in</strong>gton, UK) and run on an automated<br />
sequencer (ABI Prism 310 DNA sequencer, Applied
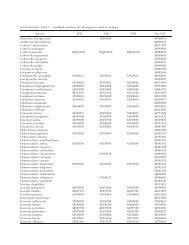
TABLE II. Primers used <strong>in</strong> this study and PCR product lengths<br />
Biosystems, Foster City, California). ITS sequences were<br />
subjected to BLAST analysis (http://www.ncbi.nlm.nih.<br />
gov/blast) and compared to the sequences available <strong>in</strong> the<br />
databases to confirm species identification.<br />
The four novel primer pairs were used for Sybr green real<br />
time PCR. Serial dilutions correspond<strong>in</strong>g to the DNA<br />
extracted from 0.1 mg to 32 ng dried <strong>mushrooms</strong> were<br />
prepared for each <strong>of</strong> the four target species, A. phalloides, L.<br />
cristata, L. brunneo<strong>in</strong>carnata and I. asterospora. These<br />
dilutions were used to test the sensitivity and the efficiency<br />
<strong>of</strong> the four protocols <strong>in</strong> triplicate. The four thermal pr<strong>of</strong>iles<br />
were identical: 95 C for 2 m<strong>in</strong>, 35 cycles at 95 C for 15 s and<br />
at 57 C for 30 s, and melt curve 55–95 C with <strong>in</strong>crements <strong>of</strong><br />
0.5 C per cycle. All reactions were performed <strong>in</strong> 25 mL, with<br />
600 nM <strong>of</strong> each primer, 12.5 mL iQ-SybrGreen Supermix<br />
and 1 mL DNA extraction. All dilutions also were tested <strong>in</strong><br />
conventional PCR for a sensitivity comparison.<br />
Subsequently all extracted DNA (TABLE III) were analyzed<br />
on real time PCR with the primers targeted to the<br />
species present <strong>in</strong> the sample. Reconstruction <strong>of</strong> possible<br />
poison<strong>in</strong>g conditions was attempted by mix<strong>in</strong>g gastric<br />
aspirates without mushroom spores with known quantities<br />
<strong>of</strong> dried <strong>mushrooms</strong> and <strong>in</strong>cubat<strong>in</strong>g them for two different<br />
time spans (i.e. 12 h and 24 h) at 36 C. PCR protocols also<br />
were tested on different samples <strong>of</strong> food conta<strong>in</strong><strong>in</strong>g<br />
<strong>poisonous</strong> <strong>mushrooms</strong> obta<strong>in</strong>ed from the Laboratorio di<br />
Sanità Pubblica <strong>of</strong> Milano (Italy).<br />
RESULTS<br />
A total <strong>of</strong> 70 DNA samples were obta<strong>in</strong>ed, 40 from 31<br />
species <strong>of</strong> both <strong>poisonous</strong> and edible fungi (TABLE I)<br />
and 30 from cl<strong>in</strong>ical samples. All samples were<br />
analyzed for DNA quantity and quality with a<br />
spectrophotometer. All samples exhibited a DNA<br />
concentration above 20 ng/mL and 260/280 ratio<br />
1.7–2.1 and thus were considered suitable for<br />
subsequent analyses. All samples were positive <strong>in</strong><br />
PCR for Basidiomycetes ITS, with the exception <strong>of</strong> the<br />
DNA samples obta<strong>in</strong>ed from gastric aspirates without<br />
<strong>mushrooms</strong>, used as negative controls.<br />
EPIS ET AL.: POISONOUS MUSHROOMS 751<br />
Primers Sequence (59–39) PCR product Amplified gene Reference<br />
ITS-Aph-F CTGTCTGCTTTTTTGATAGGTA 158 bp ITS2 This study<br />
ITS-Aph-R CAGAGAGAAGTGATATTGCTC This study<br />
ITS-Lcr-F TGACTCCTCGAACGGCTT 106 bp ITS1 This study<br />
ITS-Lcr-R TGGAAAAGACATAGACCTAG This study<br />
ITS-Lbr-F CATGCTGGCTTTGTAAGG 125 bp ITS2 This study<br />
ITS-Lbr-R ATTATCACACCGGCAACTGA This study<br />
ITS-Ias-F ATATGATGTGGCTTTTGGATGATG 94 bp ITS2 This study<br />
ITS-Ias-R AGTAGCCCCTCAGATACCA This study<br />
ITS1 TCCGTAGGTGAACCTGCGG Variable ITS1 Withe et al. 1990<br />
ITS2 GCTGCGTTCTTCATCGATGC Withe et al. 1990<br />
ITS3 GCATCGATGAAGAACGCAGC Variable ITS2 Withe et al. 1990<br />
ITS4 TCCTCCGCTTATTGATATGC Withe et al. 1990<br />
All these samples were used to test the specificity<br />
and sensitivity <strong>of</strong> each <strong>of</strong> the primers pairs designed<br />
for real time PCR. All protocols were specific (i.e. the<br />
amplification was obta<strong>in</strong>ed only when DNA from the<br />
target species was present), with no aspecific amplification<br />
observed on agarose gel or <strong>in</strong> real time PCR.<br />
The amplified fragments were sequenced, and the<br />
sequences matched those deposited <strong>in</strong> databanks for<br />
each <strong>of</strong> the four species.<br />
The maximum sensitivity and the efficiency for the<br />
four real time PCR protocols are <strong>in</strong>dicated (respectively<br />
<strong>in</strong> TABLE III and FIG. 1). Conventional PCR<br />
amplification <strong>of</strong> mushroom DNA <strong>in</strong> serial dilution<br />
also was performed on all samples with the same<br />
primers pairs and thermal pr<strong>of</strong>ile as for the real time<br />
PCR. Conventional PCR exhibited sensitivity 10–100<br />
times lower than real time PCR (data not shown).<br />
Fragmented dried <strong>mushrooms</strong> were added to<br />
aliquots <strong>of</strong> gastric juice to simulate the samples that<br />
typically are obta<strong>in</strong>ed from poisoned patients. These<br />
samples were subjected to DNA extraction and real<br />
time PCR to evaluate the sensitivity <strong>of</strong> the protocols<br />
under conditions similar to those encountered <strong>in</strong><br />
diagnoses. Real time PCR amplification <strong>of</strong> the<br />
samples treated with gastric juice showed Ct (cycle<br />
threshold) values that were actually slightly higher<br />
than those obta<strong>in</strong>ed from samples not treated with<br />
gastric juice (a portion <strong>of</strong> the results are shown <strong>in</strong><br />
TABLE III). For example the DNA sample extracted<br />
from 10 mL aspirate with addition <strong>of</strong> 1 mg dried A.<br />
phalloides kept 24 h at 36 C presents a mean Ct value<br />
<strong>of</strong> 20.45, which is close to the 19.53 Ct obta<strong>in</strong>ed from<br />
4 mg dried mushroom.<br />
DISCUSSION<br />
Mushrooms from species A. phalloides, L. cristata, L.<br />
brunneo<strong>in</strong>carnata and I. asterospora can be identified<br />
mistakenly as edible by the collector, thus possibly
752 MYCOLOGIA<br />
TABLE III. Samples exam<strong>in</strong>ed by real time PCR and ma<strong>in</strong> results <strong>of</strong> the study<br />
Target species Samples Start<strong>in</strong>g material Mean cycle-threshold Standard deviation<br />
A. phalloides A. phalloides dried mushroom 0.1 mg 12.72 0.07<br />
A. phalloides A. phalloides dried mushroom 32 ng 31.00 0.15<br />
A. phalloides 10 mL Aspirate with addition <strong>of</strong> 5 mg dried mushroom 0.05 mg 17.58 0.51<br />
A. phalloides Aspirate without addition <strong>of</strong> <strong>mushrooms</strong> 0 mg N/A N/A<br />
A. phalloides 10 mL Aspirate with addition 10 mg dried mushroom Sample 1 0.1 mg 12.87 0.18<br />
A. phalloides 10 mL Aspirate with addition 10 mg dried mushroom Sample 2 0.1 mg 12.98 0.12<br />
A. phalloides 10 mL Aspirate with addition 10 mg dried mushroom Sample 3 0.1 mg 12.76 0.13<br />
A. phalloides 10 mL Aspirate with addition 10 mg dried mushroom Sample 4 0.1 mg 13.90 0.14<br />
A. phalloides 10 mL Aspirate with addition 10 mg dried mushroom Sample 5 0.1 mg 12.76 0.08<br />
A. phalloides 10 mL Aspirate with addition 1 mg dried mushroom 12 h at 36 C 0.01 mg 17.89 0.11<br />
A. phalloides 10 mL Aspirate with addition 1 mg dried mushroom 24 h at 36 C 0.01 mg 20.45 0.20<br />
A. phalloides 10 mL aspirate at 36 C 12 h without mushroom 0 mg N/A N/A<br />
A. phalloides Mixed cooked <strong>mushrooms</strong> sample 1 0.2 mg 18.50 0.11<br />
A. phalloides Mixed cooked <strong>mushrooms</strong> sample 2 0.2 mg 20.11 0.06<br />
A. phalloides Mixed cooked <strong>mushrooms</strong> sample 3 0.2 mg 18.02 0.05<br />
A. phalloides Pasta with <strong>mushrooms</strong> 0.2 mg 17.04 0.11<br />
L. cristata L. cristata dried <strong>mushrooms</strong> 0.1 mg 17.96 0.062<br />
L. cristata L. cristata dried <strong>mushrooms</strong> 32 ng 29.80 0.158<br />
L. cristata 10 mL aspirate with addition 1 mg dried mushroom 12 h at 36 C 0.01 mg 24.57 0.09<br />
L. cristata 10 mL aspirate with addition 1 mg dried mushroom 24 h at 36 C 0.01 mg 25.79 0.41<br />
L. cristata 10 mL aspirate at 36 C 12 h without mushroom 0 mg N/A N/A<br />
L. cristata Mixed cooked <strong>mushrooms</strong> 0.2 mg 26.71 0.19<br />
L. cristata Pasta with <strong>mushrooms</strong> 0.2 mg 27.06 0.50<br />
L. brunneo<strong>in</strong>carnata L. brunneo<strong>in</strong>carnata dried mushroom 0.1 mg 14.73 0.03<br />
L. brunneo<strong>in</strong>carnata L. brunneo<strong>in</strong>carnata dried mushroom 32 ng 27.57 0.08<br />
L. brunneo<strong>in</strong>carnata 10 mL aspirate with addition 5 mg dried mushroom Sample 1 0.05 mg 24.91 0.12<br />
L. brunneo<strong>in</strong>carnata 10 mL aspirate with addition 5 mg dried mushroom Sample 2 0.05 mg 25.24 0.05<br />
L. brunneo<strong>in</strong>carnata 10 mL aspirate with addition 1 mg dried mushroom 12 h at 36 C 0.01 mg 25.17 0.11<br />
L. brunneo<strong>in</strong>carnata 10 mL aspirate with addition 1 mg dried mushroom 24 h at 36 C 0.01 mg 26.72 0.12<br />
L. brunneo<strong>in</strong>carnata 10 mL aspirate at 36 C 12 h without mushroom 0 mg N/A N/A<br />
L. brunneo<strong>in</strong>carnata Mixed cooked <strong>mushrooms</strong> 0.2 mg 26.05 0.35<br />
I. asterospora I. asterospora dried mushroom 0.1 mg 12.39 0.095<br />
I. asterospora I. asterospora dried mushroom 32 ng 25.03 0.03<br />
I. asterospora 10 mL aspirate with addition 1 mg dried mushroom 12 h at 36 C 0.01 mg 19.02 0.26<br />
I. asterospora 10 mL aspirate with addition 1 mg dried mushroom 24 h at 36 C 0.01 mg 22.06 0.02<br />
I. asterospora 10 mL aspirate at 36 C 12 h without mushroom 0 mg N/A N/A<br />
I. asterospora 10 mL aspirate with 5 mg dried mushroom 0.05 mg 24.98 0.07<br />
I. asterospora Pasta with <strong>mushrooms</strong> 0.2 mg 15.20 0.01
caus<strong>in</strong>g poison<strong>in</strong>gs. In cases <strong>of</strong> suspected mushroom<br />
poison<strong>in</strong>g species identification based on morphologic<br />
characters is <strong>of</strong>ten difficult; the morphology <strong>of</strong><br />
the <strong>mushrooms</strong>, particularly <strong>of</strong> the spores, may be<br />
distorted by handl<strong>in</strong>g and cook<strong>in</strong>g, and a mycologist<br />
might be unable to identify the species. Exam<strong>in</strong>ation<br />
<strong>of</strong> fungal spores <strong>in</strong> the gastric contents also may be<br />
<strong>in</strong>conclusive. If poison<strong>in</strong>g by A. phalloides-type <strong>mushrooms</strong><br />
is suspected, gastric contents, mushroom<br />
samples and residuals <strong>of</strong> food if available must be<br />
assayed to verify the presence <strong>of</strong> <strong>mushrooms</strong> or<br />
spores. The development <strong>of</strong> methods for the identification<br />
<strong>of</strong> <strong>poisonous</strong> <strong>mushrooms</strong> thus is important.<br />
Maeta et al. (2008) present such a methodology for<br />
four <strong>poisonous</strong> species common <strong>in</strong> Japan.<br />
As for the four mushroom species for which we<br />
developed the real time PCR, a molecular method for<br />
<strong>detection</strong> so far has been published only for A.<br />
phalloides (Kotlowski et al. 2000). This method was<br />
based on a conventional PCR. It must be emphasized<br />
EPIS ET AL.: POISONOUS MUSHROOMS 753<br />
FIG. 1. Serial dilution <strong>of</strong> each dried mushroom species. Amanita phalloides. A. Efficiency <strong>of</strong> the reaction: 101.1%.B.Inocybe<br />
asterospora. Efficiency <strong>of</strong> the reaction: 93.3%.C.Lepiota cristata. Efficiency <strong>of</strong> the reaction: 98.7%.D.Lepiota brunneo<strong>in</strong>carnata.<br />
Efficiency <strong>of</strong> the reaction: 98.6%.<br />
that the <strong>detection</strong> <strong>of</strong> a specific fungus requires a few<br />
hours with conventional PCR, while real time PCR<br />
requires only 1 h or less depend<strong>in</strong>g on the apparatus.<br />
In addition Kotlowski et al. did not present an<br />
application to cl<strong>in</strong>ical samples.<br />
Here we present a real time PCR protocol for the<br />
<strong>detection</strong> <strong>of</strong> four medically important <strong>poisonous</strong><br />
mushroom species. All protocols were highly specific<br />
for the target species and sufficiently sensitive to<br />
detect up to 32 ng dried mushroom. Furthermore, as<br />
demonstrated by Maeta et al. (2008), fungal DNA is<br />
detectable <strong>in</strong> various cooked preparations. We focused<br />
on samples that are particularly difficult for<br />
morphological identification, such as pasta with<br />
<strong>mushrooms</strong> (TABLE III), where the action <strong>of</strong> starch<br />
on the spores makes morphological details <strong>in</strong>dist<strong>in</strong>guishable<br />
while the DNA is expected to rema<strong>in</strong> <strong>in</strong><br />
sufficiently good condition.<br />
The real time PCR amplification <strong>of</strong> the samples <strong>of</strong><br />
the four species <strong>of</strong> <strong>mushrooms</strong> treated with gastric
754 MYCOLOGIA<br />
juice showed higher Ct values than untreated ones.<br />
For example the DNA sample extracted from 10 mL<br />
aspirate with addition <strong>of</strong> 1 mg dried A. phalloides kept<br />
24 h at 36 C (start<strong>in</strong>g material 10 mg) presents a mean<br />
Ct value <strong>of</strong> 20.45 while the sample not treated with<br />
the gastric aspirate conta<strong>in</strong><strong>in</strong>g a similar start<strong>in</strong>g<br />
quantity <strong>of</strong> mushroom (4 mg) had a Ct <strong>of</strong> 19.53 (data<br />
not shown). In any case the protocols also exhibited<br />
high specificity and sensitivity on the samples treated<br />
with gastric juice. The fact that the Ct values from<br />
these samples were not too divergent from the ones<br />
generated from dried specimens suggests that treatment<br />
with gastric juice lead only to a moderate<br />
degradation <strong>of</strong> the mushroom DNA.<br />
It must be emphasized however that some treatments<br />
can <strong>in</strong>fluence different samples <strong>in</strong> different<br />
ways and the result might not always be predictable.<br />
For example for I. asterospora the DNA sample<br />
obta<strong>in</strong>ed from 10 mL aspirate with addition <strong>of</strong> 1 mg<br />
dried mushroom for 24 h at 36 C shows a lower Ct<br />
value than the sample obta<strong>in</strong>ed from 10 mL aspirate<br />
with 5 mg dried mushroom while <strong>in</strong> the case <strong>of</strong> A.<br />
phalloides the DNA sample obta<strong>in</strong>ed from 10 mL<br />
aspirate with 5 mg dried mushroom shows a lower Ct.<br />
A possible explanation is a difference between the<br />
start<strong>in</strong>g samples, which could conta<strong>in</strong> DNA-degrad<strong>in</strong>g<br />
substances or PCR <strong>in</strong>hibitors, as well as a difference <strong>in</strong><br />
the gastric aspirates collected from different patients<br />
and thus likely vary<strong>in</strong>g <strong>in</strong> pH, enzyme content, etc.<br />
In conclusion the real time PCR protocol presented<br />
here exhibits a number <strong>of</strong> features that make it a<br />
useful diagnostic tool. It is specific, sensitive, quick,<br />
relatively cheap and can function with samples that<br />
are difficult to identify morphologically. Future<br />
developments <strong>of</strong> this technique could <strong>in</strong>clude novel<br />
primers for other <strong>poisonous</strong> mushroom species and<br />
the implementation <strong>of</strong> a multiplex real time PCR<br />
protocol to test <strong>in</strong> a s<strong>in</strong>gle analysis a cl<strong>in</strong>ical sample<br />
for the presence <strong>of</strong> different fungal DNA.<br />
ACKNOWLEDGMENTS<br />
The authors thank Dr Massimo Pajoro for help <strong>in</strong> DNA<br />
extraction procedures and Gruppo Micologico ‘‘Ercole<br />
Cantù’’ Agrate Brianza for mushroom samples. The authors<br />
also thank the two anonymous reviewers for useful<br />
comments and suggestions.<br />
LITERATURE CITED<br />
Assisi F, Balestreri S, Galli R. 2008. Funghi velenosi. Italy:<br />
Dalla Natura. 368 p.<br />
Bao DP, Aimi T, Kitamoto Y. 2005. Cladistic relationships<br />
among the Pleurotus ostreatus complex, the Pleurotus<br />
pulmonarius complex, and Pleurotus eryngii based on<br />
the mitochondrial small subunit ribosomal DNA<br />
sequence analysis. J Wood Sci 51:77–82.<br />
Barbato MP. 1993. Poison<strong>in</strong>g from accidental <strong>in</strong>gestion <strong>of</strong><br />
<strong>mushrooms</strong>. Med J Aust. 158:842–847.<br />
Bon M. 1988. Champignons d’Europe occidentale. France:<br />
Arthaud. 184 p.<br />
Hall AH, Spoerke DG, Rumack BH. 1987. Mushroom<br />
poison<strong>in</strong>g: identification, diagnosis and treatment.<br />
Pediatr Rev 8:291–298.<br />
Karlson-Stiber C, Persson H. 2003. Cytotoxic fungi—an<br />
overview. Toxicon 15:339–349.<br />
Kotlowski R, Myjak P, Kur J. 2000. Specific <strong>detection</strong> <strong>of</strong><br />
Amanita phalloides mycelium and spores by PCR<br />
amplification <strong>of</strong> the gpd (glyceraldehyde-3-phosphate<br />
dehydrogenase) gene fragment. J Food Biochem 24:<br />
201–212.<br />
Lark<strong>in</strong> MA, Blackshields G, Brown NP, Chenna R, McGettigan<br />
PA, McWilliam H, Valent<strong>in</strong> F, Wallace IM, Wilm A,<br />
Lopez R, Thompson JD, Gibson TJ, Higg<strong>in</strong>s DG. 2007.<br />
Nov Clustal W and Clustal X version 2.0. Bio<strong>in</strong>formatics<br />
1:2947–2948.<br />
Lurie Y, Wasser SP, Taha M, Shehade H, Nijim J, H<strong>of</strong>fmann<br />
Y, Basis F, Vardi M, Lavon O, Suaed S, Bisharat B,<br />
Bentur Y. 2009. Mushroom poison<strong>in</strong>g from species <strong>of</strong><br />
genus Inocybe (fiber head mushroom): a case series<br />
with exact species identification. Cl<strong>in</strong> Toxicol 47:562–<br />
565.<br />
Maeta K, Ochi T, Tokimoto K, Shimomura N, Maekawa N,<br />
Kawaguchi N, Nakaya M, Kitamoto Y, Aimi T. 2008.<br />
Rapid species identification <strong>of</strong> cooked <strong>poisonous</strong><br />
<strong>mushrooms</strong> by us<strong>in</strong>g real time PCR. Appl Environ<br />
Microbiol 74:3306–3309.<br />
McPartland JM, Vilgalys RJ, Cubeta MA. 1997. Mushroom<br />
poison<strong>in</strong>g. Am Fam Physician 55:1797–1800, 1805–<br />
1809, 1811–1812.<br />
Mazza R. 1995. I funghi—Guida al riconoscimento. Italy:<br />
Manuali Sonzogno. 164 p.<br />
Moser M. 1993. Guida alla determ<strong>in</strong>azione dei funghi.<br />
Vol. 1. Polyporales, Boletales, Agaricales, Russulales<br />
Italy: Saturnia. 565 p.<br />
Palapala VA, Aimi T, Inatomi S, Mor<strong>in</strong>aga T. 2002. ITS-PCR-<br />
RFLP method for the dist<strong>in</strong>guish<strong>in</strong>g commercial<br />
cultivars <strong>of</strong> edible mushroom, Flammul<strong>in</strong>a velutipes. J<br />
Food Sci 67:2486–2490.<br />
Roux X, Labadie P, Morand C, Fonta<strong>in</strong>e B, Coutant G.<br />
2008. Mushroom poison<strong>in</strong>g by brunneo<strong>in</strong>carnata:<br />
about two cases. Ann Fr Anesth Reanim 27:450–452.<br />
Schmidt O, Moreth U. 2000. Species-specific PCR primers<br />
<strong>in</strong> the rDNA-ITS region as a diagnostic tool for Serpula<br />
lacrymans. Mycol Res 104:69–72.<br />
Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani<br />
AM. 2000. Rapid identification <strong>of</strong> fungi by us<strong>in</strong>g the<br />
ITS2 genetic region and an automated fluorescent<br />
capillary electrophoresis system. J Cl<strong>in</strong> Microbiol 38:<br />
944.<br />
White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and<br />
direct sequenc<strong>in</strong>g <strong>of</strong> fungal ribosomal RNA genes for<br />
phylogenetics. In: Innis MA, Gelfand DH, Sn<strong>in</strong>sky JJ,<br />
White TJ, eds. PCR protocols: a guide to methods and<br />
applications. New York: Academic Press. p 315–322.