Species boundaries within Saprolegnia (Saprolegniales ... - Mycologia
Species boundaries within Saprolegnia (Saprolegniales ... - Mycologia
Species boundaries within Saprolegnia (Saprolegniales ... - Mycologia
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>Mycologia</strong>, 99(3), 2007, pp. 421–429.<br />
# 2007 by The Mycological Society of America, Lawrence, KS 66044-8897<br />
<strong>Species</strong> <strong>boundaries</strong> <strong>within</strong> <strong>Saprolegnia</strong> (<strong>Saprolegnia</strong>les, Oomycota) based on<br />
morphological and DNA sequence data<br />
Jonathan P. Hulvey<br />
David E. Padgett<br />
J. Craig Bailey1 Department of Biology and Marine Biology, University<br />
of North Carolina Wilmington, 601 S. College Road,<br />
Wilmington, North Carolina 28403<br />
Abstract: <strong>Saprolegnia</strong> is a common and widespread<br />
genus of Oomycetes, however species identifications<br />
are difficult and uncertain. To test whether keys based<br />
on morphological characters could identify species as<br />
determined by molecular characters we determined<br />
partial DNA sequences for the 28S rRNA gene and<br />
the complete internal transcribed spacer (ITS) region<br />
for 55 isolates belonging to <strong>Saprolegnia</strong> and one<br />
isolate of Protoachlya hypogyna that exhibited saprolegnoid<br />
zoospore discharge in water culture. Phylogenetic<br />
analyses of the combined sequence data<br />
yielded 10 robustly supported clades that probably<br />
represent separate species. Morphological analyses of<br />
all isolates revealed that each DNA-based clade could<br />
be delimited from others by autapomorphic or<br />
unique combinations of morphological character<br />
states but not without employing several features<br />
previously not used at the species level. Taxonomic<br />
implications of these results are discussed and<br />
recommendations for less equivocal characterization<br />
of new <strong>Saprolegnia</strong> species are made.<br />
Key words: morphology, phylogeny, systematics,<br />
watermold<br />
INTRODUCTION<br />
<strong>Species</strong> of the oomycetous family <strong>Saprolegnia</strong>ceae,<br />
commonly referred to as ‘‘watermolds’’ (history of<br />
term recounted in Johnson et al 2002), occur<br />
worldwide and can be isolated from freshwater and<br />
nonsaline soils with relative ease. Representatives are<br />
most notably characterized by the production of<br />
coenocytic, filamentous thalli, biflagellate, heterokont<br />
zoospores, and oogamous sexual structures.<br />
Several genera include significant pathogens of fishes<br />
(Willoughby 1997, Paxton and Willoughby 2000),<br />
crustaceans (Westman 1991, Vennerstrom et al 1998),<br />
midge eggs (Martin 1981), mosquito larvae (Seymour<br />
1984) and at least two important agricultural plants<br />
Accepted for publication 9 January 2006.<br />
1 Corresponding author. E-mail: baileyc@uncw.edu<br />
421<br />
(Wicker et al 2001, Yokosawa et al 1988, Pilet-Nayel et<br />
al 2002, Ramirez et al 1994, Peters and Grau 2002),<br />
but most included species probably are obligate<br />
saprotrophs.<br />
Kützing (1843) formally erected the <strong>Saprolegnia</strong>ceae<br />
but considered representatives to be nonchlorophyllous<br />
algae. The first English language monograph<br />
of the family was published by Humphrey<br />
(1893) and revised by Coker (1923). Subsequently<br />
Moreau and Moreau (1935, 1938), Coker (1935),<br />
Dick (1969, 1972, 1978, 2001), Miller and Ristanović<br />
(1975), Powell and Blackwell (1998) and Padgett et al<br />
(2000) have made observations about watermold<br />
systematic concepts most of which emphasize the<br />
significant morphological plasticity <strong>within</strong> the family.<br />
Johnson et al (2002) offered an extensive systematic<br />
revision of the group recognizing 17 genera and 122<br />
species but agreed that significant species overlap<br />
persists because of this plasticity.<br />
Few investigators worldwide specialize in watermold<br />
systematics although several laboratories have presented<br />
molecular phylogenies of the family or of the<br />
class Peronosporomycetes to which it belongs (e.g.<br />
Dick 1999, Riethmueller et al 1999, Leclerc et al 2000,<br />
Peterson and Rosendahl 2000, Spencer et al 2002,<br />
Daugherty et al 1998, Hudspeth et al 2000, Inaba and<br />
Tokumasu 2002). To our knowledge however there<br />
have been no previous assessments of the degree to<br />
which gene sequence analyses might contribute to<br />
resolving the species overlap problem that has grown<br />
out of the use of morphological features as the sole<br />
basis for species circumscription.<br />
The present contribution focuses exclusively on<br />
isolates belonging to the genus <strong>Saprolegnia</strong> as determined<br />
by zoosporangial discharge pattern. Its sole<br />
objective is to determine whether all isolates of an<br />
inferred phylogenetic genetic clade belong to the<br />
same morphological species as determined by modern<br />
dichotomous keys ( Johnson et al 2002). If the<br />
trees derived from our sequence alignment are<br />
robustly supported, it seems logical to assume that<br />
all isolates belonging to the same clade should be<br />
recognized as a single (or ‘‘good’’) phylogenetic<br />
species. Thus this manuscript tests the congruency<br />
between modern molecular analyses and classical<br />
morphological identification for <strong>Saprolegnia</strong> species.<br />
Partial 28S ribosomal RNA (28S rRNA) gene<br />
sequences and complete ribosomal internal transcribed<br />
spacer (ITS) sequences were determined for
422 MYCOLOGIA<br />
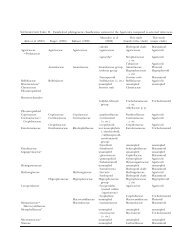
TABLE I. Source and sequence accession numbers of isolates studied. Isolates designated as ‘sp.’ reproduced only asexually<br />
under the present culture conditions. Some ATCC or CBS isolates did not key to the same species as given by the source<br />
organization. For those the original identification is indicated by * and our identification is indicated by **.<br />
UNCW<br />
No.<br />
ATCC or CBS<br />
No. <strong>Species</strong> Collection locale<br />
GenBank accession No.<br />
28S rRNA ITS<br />
105 — S. megasperma Soil, New Hanover County, NC DQ393440 DQ393506<br />
114 — <strong>Saprolegnia</strong> sp. Fish, Pamlico River, NC DQ393441 DQ393507<br />
162 — S. megasperma Carolina Biological Supply DQ393442 DQ393508<br />
170 — <strong>Saprolegnia</strong> sp. Soil, New Hanover County, NC DQ393443 DQ393509<br />
172 — <strong>Saprolegnia</strong> sp. Soil, New Hanover County, NC DQ393444 DQ393510<br />
176 — S. diclina Soil, New Hanover County, NC DQ393445 DQ393511<br />
217 — S. diclina Soil, New Hanover County, NC DQ393447 DQ393513<br />
218 — <strong>Saprolegnia</strong> sp. Soil, New Hanover County, NC DQ393448 DQ393514<br />
219 — <strong>Saprolegnia</strong> sp. Soil, New Hanover County, NC DQ393449 DQ393515<br />
250 — <strong>Saprolegnia</strong> sp. Soil, Croatan Nat. Forest, NC DQ393451 DQ393517<br />
251 — <strong>Saprolegnia</strong> sp. Soil, Croatan Nat. Forest, NC DQ393452 DQ393518<br />
253 — S. terrestris Soil, Croatan Nat. Forest, NC DQ393453 DQ393519<br />
254 — S. megasperma Soil, Croatan Nat. Forest, NC DQ393454 DQ393520<br />
257 — S. megasperma Soil, Croatan Nat. Forest, NC DQ393455 DQ393521<br />
258 — S. megasperma Soil, New Hanover County, NC DQ393456 DQ393522<br />
259 — S. richteri Soil, Great Smoky Mtns. Nat. Pk., NC DQ393457 DQ393523<br />
260 — S. luxurians Soil, Great Smoky Mtns. Nat. Pk., NC DQ393458 DQ393524<br />
263 ATCC 26116 S. ferax* S. litoralis** Pond water, California DQ393460 DQ393527<br />
264 ATCC 22284 S. parasitica* <strong>Saprolegnia</strong> sp.** Catfish, Virginia pond DQ393461 DQ393528<br />
266 ATCC 10396 S. ferax* <strong>Saprolegnia</strong> sp.** DQ393462 DQ393529<br />
269 ATCC 36146 S. ferax* <strong>Saprolegnia</strong> sp. ** Water, England DQ393463 DQ393530<br />
271 ATCC90213 S. parasitica* <strong>Saprolegnia</strong> sp. ** DQ393464 DQ393531<br />
273 ATCC 200013 S. parasitica* <strong>Saprolegnia</strong> sp. ** Fish, Japan DQ393466 DQ393532<br />
277 — <strong>Saprolegnia</strong> sp. Crayfish, Odum, GA DQ393467 DQ393533<br />
278 — <strong>Saprolegnia</strong> sp. Pet store fish, Wilmington, NC DQ393468 DQ393525<br />
280 — S. itoana Soil, Fayetteville, NC DQ393469 DQ393534<br />
283 — S. megasperma Creek bank, Wayne County, GA DQ393470 DQ393535<br />
284 — S. megasperma Water, Wayne County, GA DQ393471 DQ393536<br />
286 — S. megasperma Water, Wayne County, GA DQ393473 DQ393536<br />
288 — <strong>Saprolegnia</strong> sp. Water, Wayne County, GA DQ393474 DQ393537<br />
289 — <strong>Saprolegnia</strong> sp. Soil, Okefenokee Swamp, GA DQ393475 DQ393538<br />
290 — <strong>Saprolegnia</strong> sp. Water, Wayne County, GA DQ393476 DQ393539<br />
291 — <strong>Saprolegnia</strong> sp. Soil, Okefenokee Swamp, GA DQ393488 DQ393550<br />
292 — <strong>Saprolegnia</strong> sp. Soil, Okefenokee Swamp, GA DQ393477 DQ393540<br />
295 ATCC 36144 S. diclina* <strong>Saprolegnia</strong> sp. ** Water, England DQ393478 DQ393541<br />
298 — <strong>Saprolegnia</strong> sp. Soil, Okefenokee Swamp, GA DQ393479 DQ393542<br />
299 — <strong>Saprolegnia</strong> sp. Soil, Okefenokee Swamp, GA DQ393480 DQ393543<br />
300 — <strong>Saprolegnia</strong> sp. Soil, Italy DQ393489 DQ393551<br />
301 CBS 261.34 Pythiopsis cymosa United Kingdom DQ393490 DQ393552<br />
309 CBS 158.45 Protoachlya paradoxa* Water, Netherlands DQ393491 DQ393553<br />
312 — <strong>Saprolegnia</strong> sp. Soil, Richmond, VA DQ393481 DQ393544<br />
314 — <strong>Saprolegnia</strong> sp. Soil, Richmond, VA DQ393482 DQ393545<br />
315 — <strong>Saprolegnia</strong> sp. Soil, Richmond, VA DQ393483 DQ393546<br />
320 — S. megasperma Soil, Great Smoky Mtns. Nat. Pk., NC DQ393485 DQ393547<br />
328 — <strong>Saprolegnia</strong> sp. Pet store goldfish, Wilmington, NC DQ393486 DQ393548<br />
329 — <strong>Saprolegnia</strong> sp. Frog, Wilmington, NC DQ393487 DQ393549<br />
336 ATCC 28092 Protoachlya polysporus* Water, New Jersey DQ393492 DQ393554<br />
337 ATCC 44892 Protoachyla paradoxa* Pond water, Netherlands DQ393493 DQ393555<br />
372 CBS 551.62 S. eccentrica* S. richteri** Soil, England DQ393494 DQ393556<br />
373 — S. diclina Soil, Wilmington, NC DQ393495 DQ393557<br />
374 CBS 535.67 S. litoralis* <strong>Saprolegnia</strong> sp. ** Soil, England DQ393496 DQ393558<br />
375 CBS 213.35 S. unispora* <strong>Saprolegnia</strong> sp. ** United Kingdom DQ393497 DQ393559
TABLE I. Continued<br />
UNCW<br />
No.<br />
56 isolates referable to <strong>Saprolegnia</strong>. The combined<br />
data were analyzed yielding phylogenetic trees onto<br />
which we mapped sequence divergence values and<br />
morphological character states. Results suggest that<br />
the DNA-based phylogenetic hypothesis provides<br />
a useful framework for re-interpreting the taxonomic<br />
utility of vegetative and reproductive characters for<br />
distinguishing among <strong>Saprolegnia</strong> species. This more<br />
objective approach, in which both molecular and<br />
morphological data are employed, may provide<br />
a method for ultimately establishing nonoverlapping<br />
generic and species descriptions (and keys) for other<br />
watermold taxa as well.<br />
MATERIALS AND METHODS<br />
Culture propagation and morphological analyses.—Isolates<br />
employed in this study were derived from nonsaline<br />
soils collected principally from the southeastern United<br />
States or were purchased from the American Type<br />
Culture collection (ATCC) or the Centraalbureau voor<br />
Schimmelcultures (CBS) (TABLE I). Before morphological<br />
analysis or DNA extraction was attempted all isolates were<br />
processed to derive axenic, single spore or single hyphal tip<br />
cultures.<br />
Replicate cultures (10 for each isolate) used for morphological<br />
identifications were initiated by placing autoclaved,<br />
shelled hemp seeds on the growing edge of colonies<br />
propagated on Difco cornmeal agar (CMA). After a 24 h<br />
infestation period, single hemp seeds were transferred to<br />
disposable plastic Petri dishes filled with 25 mL of sterile<br />
distilled water. Water culture dishes were wrapped individually<br />
in Parafilm TM to prevent contamination during<br />
room temperature incubation and maintained until sacrificed<br />
to observe morphological characters. Wherever<br />
possible identifications were based on 50 replicate determinations<br />
of all morphological features studied ( Johnson et al<br />
2002).<br />
To preclude the possibility that individual primary<br />
sporangia might exhibit different patterns of sporangial<br />
dehiscence (Padgett and Johnson 2004), zoospore discharge<br />
was observed (and videotaped) from 10 separate<br />
HULVEY ET AL: SPECIES BOUNDARIES WITHIN SAPROLEGNIA 423<br />
ATCC or CBS<br />
No. <strong>Species</strong> Collection locale<br />
GenBank accession No.<br />
28S rRNA ITS<br />
377 CBS 618.97 S. polymorpha* <strong>Saprolegnia</strong> sp. ** Fish DQ393498 DQ393560<br />
380 CBS 542.67 S. furcata* <strong>Saprolegnia</strong> sp.** United Kingdom DQ393499 DQ393561<br />
381 CBS 386.52 S. megasperma * <strong>Saprolegnia</strong> sp. ** Soil, New York DQ393500 DQ393562<br />
382 CBS 109568 S. semi-hypogyna* S. luxurians** DQ393501 DQ393563<br />
383 CBS 278.52 S. subterranea* S. itoana** Mud, New York DQ393502 DQ393564<br />
384 CBS 305.37 S. ferax* <strong>Saprolegnia</strong> sp. ** France DQ393503 DQ393565<br />
345 CBS 534.67 S. ferax* <strong>Saprolegnia</strong> sp. ** Lake water, England DQ393504 DQ393566<br />
386 CBS 110064 S. torulosa* S. asterophora ** Soil, Japan DQ393505 DQ393567<br />
primary sporangia for each isolate. Discharges were<br />
observed on 24–48 h old, intact colonies floating in<br />
Parafilm-sealed culture dishes with an Olympus SZX 12<br />
stereo microscope equipped with a fiber-optic illuminator<br />
and an Hitachi CE digital camera.<br />
Subsequent to zoospore discharge determination, all<br />
replicate water cultures were individually sacrificed through<br />
time (over 14 d) for observing diagnostic sexual characters<br />
as per Johnson (1956) and Seymour (1970). These<br />
observations were made with an Olympus BX 41 phase<br />
contrast microscope and derived data recorded in specially<br />
designed MS Excel TM electronic spreadsheets (Ferner<br />
unpublished) for later analysis. Measurements of various<br />
quantitative features (e.g. zoospore cyst diameter, oogonial<br />
and oospore diameters) were made with ImagePro TM<br />
computer software.<br />
DNA isolation, PCR amplification and sequencing.—Mycelia<br />
for DNA extraction were propagated in glucose, yeast<br />
extract (GY) broth containing Difco D-glucose<br />
(10.0 g L 21 ) and Difco Yeast Extract (2.5 g L 21 ). Log phase<br />
cultures were harvested aseptically, blotted on sterile filter<br />
paper, transferred to sterile Whirl Paks TM and immediately<br />
frozen at 220 C.<br />
Frozen mycelia were ground with a mortar and pestle or<br />
disrupted via sonification in 2 3 CTAB buffer and total<br />
cellular DNA extracted as described in Bailey et al (1998). A<br />
portion of the 28S rRNA gene, including hypervariable stem<br />
and loop regions between helices C1 and D2 (Ben Ali et al<br />
1999), was amplified with primers C1 (59-ACCCGCTGATT-<br />
TAAGCAT-39) and D2 (59-TCCGTGTTTCAAGACGG-39)<br />
(Leclerc et al 2000). The entire ITS region was amplified<br />
with primers ITS1 (59-TCCGTAGGTGAACCTGCGG-39) and<br />
ITS4 (59-TCCTCCGCTTATTGATATGC-39) (White et al<br />
1990). The thermocycling profile included an initial<br />
denaturation step at 94 C for 3 min, followed by 35 cycles<br />
of denaturation at 94 C for 30 s, primer annealing at 50 C<br />
for 30 s and primer extension at 72 C for 1.5 min, a final<br />
extension step for 7 min at 72 C and a 4 C soak. Amplified<br />
products were checked for correct length and yield on 0.8%<br />
ethidium bromide-stained agarose gels and purified with<br />
the GeneClean II Kit (Qbiogene, Carlsbad, California). PCR<br />
products were sequenced on both strands with the<br />
amplification primers and BigDye Terminator cycle se-
424 MYCOLOGIA<br />
quencing kit (v. 2, Applied Biosystems, Foster City,<br />
California) according to the manufacturer’s specifications.<br />
Sequences were determined on an ABI3100 automated<br />
DNA sequencer (Applied Biosystems, Foster City, California)<br />
and electropherograms were edited and assembled<br />
with Sequence Analysis (v. 1.0.1) and Sequence Navigator<br />
(v. 3.4.1) programs (Applied Biosystems, Foster City,<br />
California).<br />
Phylogenetic analyses.—Sequences were aligned by eye in<br />
SeqApp (Gilbert 1994). The model of nucleotide substitution<br />
that best fit our data was determined with<br />
Modeltest (v. 3.06, Posada and Crandall 1998) and a general<br />
time reversible model with invariant sites and rates of<br />
substitution among sites approximated by a gamma distribution<br />
(5GTR + I + G) was selected. This substitution<br />
model and neighbor joining (NJ), parsimony (MP), and<br />
maximum likelihood (ML) algorithms were used to<br />
construct trees in PAUP (v. 4.0b10, Swofford 2001).<br />
Heuristic parsimony searches were conducted with TBR<br />
branch swapping with gaps treated as missing data and<br />
random orders of sequence addition (5100). The ML tree<br />
was inferred with the heuristic search option and random<br />
orders of sequence addition (510).<br />
Pythiopsis had been resolved as sister of <strong>Saprolegnia</strong> based<br />
on analyses of 28S rRNA + ITS data (Leclerc et al 2000) and<br />
mitochondrial cox2 gene sequences (Hudspeth et al 2000)<br />
and therefore was used to root the saprolegnialean trees.<br />
Support for nodes of the trees was assessed by calculating<br />
bootstrap proportion values (Felsenstein 1985). Bootstrap<br />
values for the NJ and MP trees were based on analyses of<br />
10 000 pseudoreplicate datasets whereas the ML values were<br />
derived from 60 replicates.<br />
RESULTS<br />
Nucleotide sequences determined in this study have<br />
been submitted to GenBank (TABLE I). The combined<br />
sequence data matrix (28S rRNA + ITS) included 1678<br />
characters. All saprolegnialean sequences analyzed<br />
differed from one another by at least one substitution;<br />
the maximum number of substitutions between any<br />
two ingroup isolates was 153. Neighbor joining<br />
resulted in a single tree whereas the MP analysis<br />
yielded 7206 equally parsimonious trees of 728 steps<br />
(CI 5 0.64, RI 5 0.86). Of 1678 total characters 278<br />
(16.6%) were found to be parsimony informative. The<br />
50% majority rule parsimony consensus tree, the NJ<br />
tree and the ML tree were consistent with one another<br />
differing only with respect to relationships inferred<br />
among some terminal taxa <strong>within</strong> clades. However the<br />
number of clades resolved, the composition of the<br />
clades and their branching order were identical in all<br />
three analyses; for these reasons only the ML tree is<br />
presented (FIG 1).<br />
<strong>Saprolegnia</strong> isolates were divided among 10 clades<br />
each supported by robust bootstrap values (FIG. 1)<br />
although the branching order among clades 1, 2, 3<br />
and 4 could not be resolved. While the bootstrap<br />
value for one clade in the ML analysis was 78%, the<br />
remaining values were above 92% and all values for<br />
the 10 clades in the distance analysis were 100%<br />
(FIG. 1). No correlation was found between the<br />
number of isolates placed in a clade and the number<br />
of pairwise substitutions observed among those<br />
isolates (r 2 520.02437, p , 0.4012). The maximum<br />
mean number of substitutions observed among<br />
isolates <strong>within</strong> any of the 10 terminal clades was 10.7<br />
(clade 4) whereas a single substitution distinguished<br />
isolates SAP280 and SAP383 (clade 6). These data<br />
(FIG. 1) together with morphological information<br />
were used to justify separate recognition of clades<br />
7 vs. 8 as well as 9 vs. 10. For example mean<br />
numbers of substitutions between members of<br />
clades 7 and 8 (520.7) are nearly twice that observed<br />
among isolates placed in the most heterogeneous<br />
terminal clade (clade 4) (510.7). Similarly the mean<br />
number of substitutions between members of<br />
clades 9 and 10 was nearly three times that observed<br />
in clade 4.<br />
Two isolates (SAP 374 and SAP 380) did not fit<br />
<strong>within</strong> other bi- or multi-isolate clades thus are<br />
assumed to represent unique taxa. In addition unique<br />
(autapomorphic) morphological characters circumscribed<br />
clades 4, 8 and 9.<br />
Inspection (TABLE II) clearly reveals that, for<br />
almost all clades, included isolates did not all key to<br />
the same morphological species when identified by<br />
the keys presented in Johnson et al (2002). Furthermore<br />
in several instances the isolates did not key to<br />
the same species as indicated by the original culture<br />
source. Thus the assumption of complete congruency<br />
between gene sequence analysis and morphological<br />
identification must be rejected. Furthermore identification<br />
of species corresponding to each genetic<br />
clade using compiled morphological characters of all<br />
clade members does not yield different names for<br />
each clade (TABLE II).<br />
We analyzed our morphological data considering<br />
not only traditional sexual characters but also some<br />
zoosporangial characters in an effort to determine<br />
whether clades could be resolved from one another<br />
with a combination of morphological features. Results<br />
of this analysis are presented (FIG 2). We found that if<br />
we first separated species by what kind of discharge<br />
papilla formed on the zoosporangia we could remove<br />
clade 8. It is noteworthy that both members of clade 8<br />
exhibited long and prominently flared zoosporangial<br />
discharge papillae (FIG. 3) that were unique to this<br />
clade. The fact that resolution of numbered clades<br />
could not be accomplished without use of this and<br />
other zoosporangial characteristics (FIG. 2) represents<br />
a new finding for this genus.
HULVEY ET AL: SPECIES BOUNDARIES WITHIN SAPROLEGNIA 425<br />
FIG. 1. Maximum likelihood phylogram depicting phylogenetic relationships inferred among 56 saprolegnoid isolates,<br />
based on combined analysis of 28S rRNA and ITS DNA sequence data. The 10 terminal clades are numbered and denoted by<br />
dots. Bootstrap values (.70%, L5 maximum likelihood, P 5 parsimony, D 5 distance) are presented to the right of each<br />
numbered clade and above corresponding internal nodes.
426 MYCOLOGIA<br />
TABLE II. Best fit identifications of sexually reproducing clade members based on sexual morphological characteristics only<br />
Clade Isolate number<br />
DISCUSSION<br />
The robust bootstrap support for all 10 clades (FIG. 1)<br />
supports the conclusion that they represent distinct<br />
species. Our finding that not all members of a given<br />
clade keyed out to the same morphological species<br />
was not unexpected. The latter finding leaves no<br />
alternative to the conclusion that <strong>Saprolegnia</strong> species<br />
cannot be circumscribed solely on the basis of sexual<br />
features.<br />
Probably our most important finding was that,<br />
when we considered the compiled morphological<br />
characteristics for an entire clade, we keyed several<br />
different clades to the same species (TABLE II).<br />
Identifications based on sexual and zoosporangial<br />
characters that were unique to each clade did permit<br />
resolution to species (FIG. 1). This represents a new<br />
finding that should mandate use of unique features<br />
for circumscription not only of newly proposed<br />
species but also recircumscription of currently recognized<br />
species.<br />
This conclusion, that species can be identified only<br />
of isolate<br />
Best fit morphological identification<br />
Based on compiled characteristics of<br />
all clade isolates<br />
1 263 S. litoralis S. megasperma<br />
373 S. diclina<br />
2 283 S. megasperma S. ferax<br />
295 S. asterophora<br />
253 S. terrestris S. megasperma<br />
3 269 S. diclina<br />
284 S. megasperma<br />
105 S. megasperma<br />
162 S. megasperma<br />
254 S. megasperma<br />
251 S. megasperma<br />
176 S. diclina<br />
286 S. megasperma<br />
4 320 S. megasperma S. megasperma<br />
5 217 S. diclina S. diclina<br />
6 280 S. itoana S. itoana<br />
383 S. itoana<br />
257 S. megasperma S. megasperma<br />
7 258 S. megasperma<br />
386 S. asterophora<br />
8 260 S. luxurians S. richteri<br />
259 S. richteri<br />
9 372 S. richteri S. richteri<br />
382 S. luxurians<br />
292 S. megasperma S. megasperma<br />
289 ?<br />
10 291 S. subeccentrica<br />
277 S. megasperma<br />
using a combination of unique sexual and zoosporangial<br />
characters, agrees with Dick’s (1973) contention<br />
that a revised multivariate analytical approach<br />
likely would be needed for meaningful systematic<br />
revision of the watermolds. Indeed he argued that<br />
knowledge of the family could advance only after this<br />
is done.<br />
To aid this daunting task of systematic revision we<br />
offer these suggestions: (i) <strong>Species</strong> circumscription<br />
for <strong>Saprolegnia</strong> can be done best using the compiled<br />
sexual and zoosporangial features for all members of<br />
a clade. This can be done however only after clade<br />
membership is determined on the basis of comparative<br />
sequence analysis (i.e. DNA data for new isolates<br />
first must be compared with those previously deposited<br />
in GenBank [e.g. TABLE I]). This conclusion<br />
necessarily implies that the type species of <strong>Saprolegnia</strong><br />
taxa should be redefined and reference to DNA<br />
sequence data should be included in the new<br />
diagnoses (Bailey et al 1998, Deane et al 1998,<br />
Guillou et al 1999, Andersen et al 2002, Moro et al<br />
2002, Hoef-Emden and Melkonian 2003). With this
FIG. 2. Key based on morphological characters to<br />
phylogenetic clades <strong>within</strong> <strong>Saprolegnia</strong>.<br />
method any new or previously studied isolate can be<br />
described or identified to species regardless of<br />
whether that particular isolate produces characteristic<br />
zoosporangial or sexual features (Lange et al 2002,<br />
Hoef-Emden and Melkonian 2003, Marin et al 2003).<br />
(ii) Whereas replicate character state determinations<br />
for sexual features of <strong>Saprolegnia</strong> isolates often are<br />
not normally distributed (our unpublished data),<br />
sample sizes must be sufficiently large for each<br />
taxonomically significant character (we suggest n 5<br />
50) to render dependable isolate characterization<br />
before new taxa are described. (iii) Whereas historically<br />
descriptions of <strong>Saprolegnia</strong> species have employed<br />
ambiguous terms such as ‘‘predominant’,<br />
‘‘frequent’’, ‘‘occasional’’ and ‘‘rare’’, we propose<br />
that all listing of sexual and zoosporangial features of<br />
a proposed taxon be made using percentage occurrence<br />
values for each morphological feature based on<br />
a minimum of n 5 50 replicate determinations. (iv)<br />
Our analyses of other watermold genera (unpublished)<br />
indicate morphological plasticity equal to or<br />
greater than that presented here for <strong>Saprolegnia</strong>. We<br />
suggest therefore that these guidelines be used for<br />
revising all genera of the family <strong>Saprolegnia</strong>ceae.<br />
Results (TABLE II) suggest to us that assignment of<br />
unequivocal specific epithets to phylogenetic clades at<br />
HULVEY ET AL: SPECIES BOUNDARIES WITHIN SAPROLEGNIA 427<br />
FIG. 3. Micrographs of typical sporangial discharge<br />
papillae. a, b. Prominent, long and flared papillae (arrows)<br />
unique to clade 8. c, d. Typical papillae of other clades.
428 MYCOLOGIA<br />
this date would be unwise. The extreme divergence of<br />
our findings from classically accepted systematic<br />
criteria for <strong>Saprolegnia</strong> suggests that taxonomic<br />
criteria for this genus, and probably all others<br />
belonging to the <strong>Saprolegnia</strong>ceae, must be comprehensively<br />
reevaluated. For these reasons we have<br />
elected not to devise any temporary scheme of<br />
classification at this time.<br />
ACKNOWLEDGMENTS<br />
We gratefully acknowledge financial support provided by<br />
the National Science Foundation grant DEB 0328316<br />
(PEET program). Raymond R. Stone, Maris Durako and<br />
Zhuoer Lin provided assistance with morphological characterizations,<br />
and Melissa Smith assisted with preparation of<br />
FIG. 3. Dr T.W. Johnson Jr. (Duke Univ., retired) kindly<br />
provided advice on the morphological identifications, and<br />
critical comments on the manuscript.<br />
LITERATURE CITED<br />
Andersen RA, Potter D, Bailey JC. 2002. Pinguiococcus<br />
pyrenoidosus gen. et sp. nov. (Pinguiophyceae), a new<br />
marine coccoid alga. Phycol Res 50:57–65.<br />
Bailey JC, Bidigare RR, Christensen SJ, Andersen RA. 1998.<br />
Phaeothamniophyceae classis nova: a new lineage of<br />
chromophytes based upon photosynthetic pigments,<br />
rbcL, sequence analysis and ultrastructure. Protist 149:<br />
245–263.<br />
Ben Ali A, Wuyte J, de Wachter R, Meyer A, van de Peer Y.<br />
1999. Contribution of a variability map for eukaryotic<br />
large subunit ribosomal RNA. Nucleic Acid Res 27:<br />
2825–2831.<br />
Coker WC. 1923. The <strong>Saprolegnia</strong>ceae with notes on other<br />
water molds. Chapel Hill: University of North Carolina<br />
Press. 201 p.<br />
———. 1935. Interrelationships of the <strong>Saprolegnia</strong>les. Proc<br />
6th Int Bot Congr Amsterdam (Zesde Internationaal<br />
Botanisch Congres) 1:268–270.<br />
Daugherty J, Evans TM, Skillom T, Watson LE, Money NP.<br />
1998. Evolution of spore release mechanisms in the<br />
<strong>Saprolegnia</strong>ceae (Oomycetes): evidence from a phylogenetic<br />
analysis of internal transcribed spacer sequences.<br />
Fungal Genet Biol 24:354–363.<br />
Deane JA, Hill DR, Brett SJ, McFadden GI. 1998. Hanusia<br />
phi gen. et sp. nov. (Cryptophyceae): characterization<br />
of ‘Cryptomonas sp’. Eur J Phycol 33:149–154.<br />
Dick MW. 1969. Morphology and taxonomy of the<br />
Oomycetes, with special reference to <strong>Saprolegnia</strong>ceae,<br />
Leptomitaceae and Pythiaceae I. Sexual reproduction.<br />
New Phytol 68:751–775.<br />
———. 1972. Morphology and taxonomy of the Oomycetes,<br />
with special reference to <strong>Saprolegnia</strong>ceae, Leptomitaceae<br />
and Pythiaceae II. Cytogenetic systems. New<br />
Phytol 71:1151–1159.<br />
———. 1973. <strong>Saprolegnia</strong>les. In: Ainsworth GC, et al, eds.<br />
The Fungi, an advanced treatise. Vol. IVB. New York:<br />
Academic Press. p 113–144.<br />
———. 1978. Systematics, taxonomy and phylogeny in the<br />
Oomycetes. In: Subramanian CV, ed. Taxonomy of<br />
Fungi. Proceedings of the International Symposium on<br />
taxonomy of Fungi. Madras, India: University of<br />
Madras. p 82–87.<br />
———. 2001. Stramenipilous fungi: systematics of the<br />
Peronosporomycetes including accounts of the marine<br />
straminipilous protists, the Plasmodiophorids and<br />
similar organisms. Dordrecht: Kluwer Academic Publishers.<br />
670 p.<br />
———, Vick MC, Gibbings JG, Hedderson TA, Lopez-Lastra<br />
CC. 1999. 18S rDNA for species of Leptolegnia and<br />
other Peronosporomycetes: justification for the subclass<br />
taxa Saprolegniomycetidae and Peronosporomycetidae<br />
and division of the <strong>Saprolegnia</strong>ceae sensu lato<br />
into the Leptolegniaceae and <strong>Saprolegnia</strong>ceae. Mycol<br />
Res 103:1119–1125.<br />
Felsenstein J. 1985. Confidence limits on phylogenies: an<br />
approach using the bootstrap. Evolution 39:783–791.<br />
Gilbert DG. 1994. SeqApp: a biosequence editor and<br />
analysis program. Published on the Internet. Available<br />
at ftp.bio.indiana.edu<br />
Guillou L, Chretiennot-Dinet M-J, Medlin LK, Claustre H,<br />
Goer SL, Vaulot D. 1999. Bolidomonas: a new genus<br />
with two species belonging to a new algal class, the<br />
Bolidophyceae (Heterokonta). J Phycol 35:368–381.<br />
Hoef-Emden K, Melkonian M. 2003. Revision of the genus<br />
Cryptomonas (Cryptophyceae): a combination of molecular<br />
phylogeny and morphology provides insights<br />
into a long-hidden dimorphism. Protist 154:371–409.<br />
Hudspeth DSS, Nadler SA, Hudspeth MES. 2000. A COX2<br />
molecular phylogeny of the Peronosporomycetes.<br />
<strong>Mycologia</strong> 92:674–684.<br />
Humphrey JE. 1893. The <strong>Saprolegnia</strong>ceae of the United<br />
States, with notes on other species. Trans Am Phil Soc<br />
(NS) 17:63–148.<br />
Inaba S, Tokumasu S. 2002. Phylogenetic relationships<br />
between the genus <strong>Saprolegnia</strong> and related genera<br />
inferred from ITS sequences. 7th International Mycological<br />
Congress. Oslo, Norway. (Abstract 687)<br />
Johnson TW Jr. 1956. The genus Achlya. Ann Arbor:<br />
University of Michigan Press. 180 p.<br />
———, Seymour RL, Padgett DE. 2002. Biology and<br />
systematics of the <strong>Saprolegnia</strong>ceae. On-line publication<br />
accessible at http://dl.uncw.edu/digilib/biology/<br />
fungi/taxonomy%20and%20systematics/padgett%20<br />
book/<br />
Kützing FT. 1843. Phycologia generalis oder Anatomie,<br />
Physiologie und Systemkunde der Tange. Leipzig: F.A.<br />
Brockhaus. 458 p.<br />
Lange M, Chen Y-Q, Medlin LK. 2002. Molecular genetic<br />
delineation of Phaeocystis species (Prymnesiophyceae)<br />
using coding and non-coding regions of nuclear and<br />
plasmid genomes. Eur J Phycol 37:77–92.<br />
Leclerc MC, Guillot J, Deville M. 2000. Taxonomic and<br />
phylogenetic analysis of <strong>Saprolegnia</strong>ceae (Oomycetes)<br />
inferred from LSU rDNA and ITS sequence comparisons.<br />
Antonie van Leeuwenhoek 77:369–377.<br />
Marin B, Palm A, Klingberg M, Melkonian M. 2003.<br />
Phylogeny and taxonomic revision of plastid-contain-
ing Euglenophytes based on SSU rDNA sequence<br />
comparisons and synapomorphic signatures in the<br />
SSU rRNA secondary structure. Protist 154:99–145.<br />
Martin WW. 1981. Couchia circumplexa, a water mold<br />
parasitic in midge eggs. <strong>Mycologia</strong> 73:1143–157.<br />
Miller CE, Ristanović B. 1975. Systematic studies on some<br />
taxa of the <strong>Saprolegnia</strong>ceae. Milrobiologija, Acta Biol<br />
Iugoslavica 12:47–58.<br />
Moreau F, MoreauMme.. 1935. Remarques sur la systématique<br />
des Saprolégniacées. Bull Soc Bot France 82:354–<br />
358.<br />
———, ———. 1938. Recherches sur les Saprolégniées.<br />
Ann Sci Nat Bot (11 e sér) 1:221–358.<br />
Moro I, La Rocca N, Dalla Valle L, Pschin E, Negrisolo E,<br />
Andreoli C. 2002. Pyramimonas australis sp. now.<br />
(Prasinophyceae, Chlorophyta) from Antarctica: fine<br />
structure and molecular phylogeny. Eur J Phycol 37:<br />
103–114.<br />
Padgett DE, Johnson TW Jr. 2004. Zoosporangial discharge<br />
in a Protoachlya hypogyna (<strong>Saprolegnia</strong>ceae) isolate<br />
from southeastern North Carolina. <strong>Mycologia</strong> 96:205–<br />
207.<br />
———, Puckett JC, Strickland WM. 2000. Taxonomic<br />
implications of unusual zoospore discharge in an<br />
isolate of <strong>Saprolegnia</strong> unispora. Mycotaxon 76:171–174.<br />
Paxton CGM, Willoughby LG. 2000. Resistance of perch<br />
eggs to attack by aquatic fungi. J Fish Biol 57:562–570.<br />
Peters RD, Grau CR. 2002. Inoculation with nonpathogenic<br />
Fusarium solani increases severity of pea root rot<br />
caused by Aphanomyces euteiches. Plant Dis 86:411–414.<br />
Petersen AB, Rosendahl S. 2000. Phylogeny of the Peronosporomycetes<br />
(Oomycota) based on partial sequences<br />
of the large ribosomal subunit (LSU rDNA).<br />
Mycol Res 104:1295–1303.<br />
Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM,<br />
Baranger A, Coyne CJ. 2002. Quantitative trait loci for<br />
partial resistance to Aphanomyces root rot in pea.<br />
Theoretical Appl Genet 106:28–39.<br />
Posada D, Crandall KA. 1998. Modeltest: testing the model<br />
of DNA substitution. Bioinformatics 14:817–818.<br />
HULVEY ET AL: SPECIES BOUNDARIES WITHIN SAPROLEGNIA 429<br />
Powell MJ, Blackwell WH. 1998. Phenetic analysis of genera<br />
of the <strong>Saprolegnia</strong>ceae (Oomycetes). Mycotaxon 68:<br />
505–516.<br />
Ramirez MC, Raposo R, Mateo-Sagasta E. 1994. Occurrence<br />
of Aphanomyces cochlioides damping-off of sugar beet in<br />
Spain. Plant Dis 78:102.<br />
Riethmueller A, Weiss M, Oberwinkler F. 1999. Phylogenetic<br />
studies of Saprolegniomycetidae and related groups<br />
based on nuclear large subunit ribosomal DNA<br />
sequences. Can J Bot 77:1790–1800.<br />
Seymour RL. 1970. The genus <strong>Saprolegnia</strong>. Nov Hedwig 19:<br />
IV–124.<br />
———. 1984. Leptolegnia chapmanii, an oomycete pathogen<br />
of mosquito larvae. <strong>Mycologia</strong> 76:670–674.<br />
Spencer MA, Vick MC, Dick MW. 2002. Revision of<br />
Aplanopsis, Pythiopsis, and ‘subcentric’ Achlya species<br />
(<strong>Saprolegnia</strong>ceae) using 18S rDNA and morphological<br />
data. Mycol Res 106:549–560.<br />
Swofford DL. 2001. PAUP*: phylogenetic analysis using<br />
parsimony (*and other methods). Version 4. Sunderland,<br />
Massachusetts: Sinauer Associates.<br />
Vennerstrom P, Soderhall K, Cerenius L. 1998. The origin<br />
of two crayfish plague (Aphanomyces astaci) epizootics<br />
in Finland on noble crayfish, Astacus astacus. Ann<br />
Zoolog Fennici 35:43–46.<br />
Westman K. 1991. The crayfish fishery in Finland its past<br />
present and future. Finnish Fish Res 12:187–216.<br />
White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and<br />
direct sequencing of fungal ribosomal RNA gene for<br />
phylogenetics. In: Innis., et al, eds. PCR Protocols. San<br />
Diego: Academic Press. p 315–322.<br />
Wicker E, Hulle M, Rouxel F. 2001. Pathogenic characteristics<br />
of isolates of Aphanomyces euteiches from pea in<br />
France. Plant Pathol (Oxford) 50:433–442.<br />
Willoughby LG. 1997. Achlya diffusa (Fungi, Oomycota)<br />
from fish ponds in Thailand. Nov Hedwig 64:467–471.<br />
Yokosawa R, Sekizaki H, Kuninaga S. 1988. Attractants of<br />
Aphanomyces cochlioides zoospores contained in sugar<br />
beet seedlings. Ann Phytopathol Soc Japan 54:133–140.