PCR targeting system in Streptomyces coelicolor ... - Streptomyces UK
PCR targeting system in Streptomyces coelicolor ... - Streptomyces UK
PCR targeting system in Streptomyces coelicolor ... - Streptomyces UK
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
(E. coli Genetic Stock Center CGSC Stra<strong>in</strong> # 7637) was modified by replac<strong>in</strong>g the<br />
ampicill<strong>in</strong> resistance gene bla with the chloramphenicol resistance gene cat,<br />
generat<strong>in</strong>g pIJ790, to permit selection <strong>in</strong> the presence of Supercos1-derived cosmids<br />
(ampicill<strong>in</strong> and kanamyc<strong>in</strong> resistance).<br />
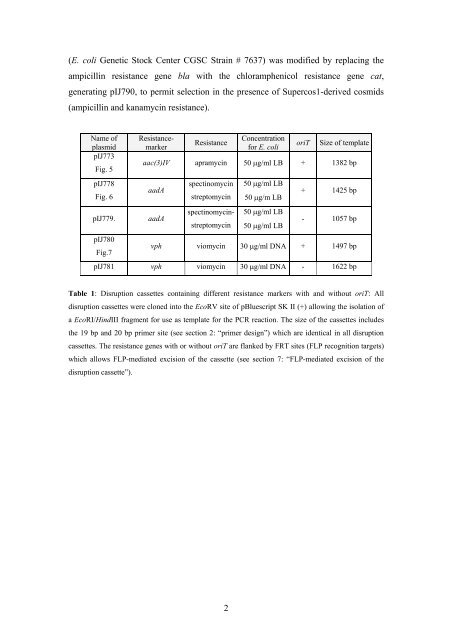
Name of<br />
plasmid<br />
pIJ773<br />
Fig. 5<br />
pIJ778<br />
Fig. 6<br />
Resistancemarker<br />
Resistance<br />
Concentration<br />
for E. coli<br />
oriT Size of template<br />
aac(3)IV apramyc<strong>in</strong> 50 μg/ml LB + 1382 bp<br />
aadA<br />
pIJ779. aadA<br />
pIJ780<br />
Fig.7<br />
spect<strong>in</strong>omyc<strong>in</strong><br />
streptomyc<strong>in</strong><br />
spect<strong>in</strong>omyc<strong>in</strong>streptomyc<strong>in</strong><br />
50 μg/ml LB<br />
50 μg/m LB<br />
50 μg/ml LB<br />
50 μg/ml LB<br />
+ 1425 bp<br />
- 1057 bp<br />
vph viomyc<strong>in</strong> 30 μg/ml DNA + 1497 bp<br />
pIJ781 vph viomyc<strong>in</strong> 30 μg/ml DNA - 1622 bp<br />
Table 1: Disruption cassettes conta<strong>in</strong><strong>in</strong>g different resistance markers with and without oriT: All<br />
disruption cassettes were cloned <strong>in</strong>to the EcoRV site of pBluescript SK II (+) allow<strong>in</strong>g the isolation of<br />
a EcoRI/H<strong>in</strong>dIII fragment for use as template for the <strong>PCR</strong> reaction. The size of the cassettes <strong>in</strong>cludes<br />
the 19 bp and 20 bp primer site (see section 2: “primer design”) which are identical <strong>in</strong> all disruption<br />
cassettes. The resistance genes with or without oriT are flanked by FRT sites (FLP recognition targets)<br />
which allows FLP-mediated excision of the cassette (see section 7: “FLP-mediated excision of the<br />
disruption cassette”).<br />
2