Prime User Manual - ISP
Prime User Manual - ISP
Prime User Manual - ISP
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
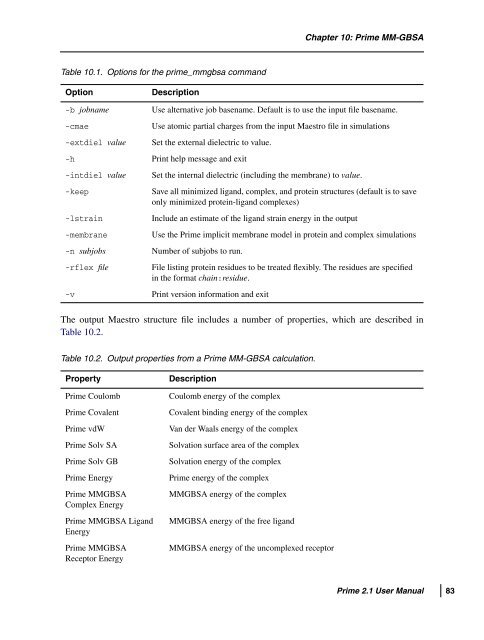
Table 10.1. Options for the prime_mmgbsa command<br />
Option Description<br />
Chapter 10: <strong>Prime</strong> MM-GBSA<br />
-b jobname Use alternative job basename. Default is to use the input file basename.<br />
-cmae Use atomic partial charges from the input Maestro file in simulations<br />
-extdiel value Set the external dielectric to value.<br />
-h Print help message and exit<br />
-intdiel value Set the internal dielectric (including the membrane) to value.<br />
-keep Save all minimized ligand, complex, and protein structures (default is to save<br />
only minimized protein-ligand complexes)<br />
-lstrain Include an estimate of the ligand strain energy in the output<br />
-membrane Use the <strong>Prime</strong> implicit membrane model in protein and complex simulations<br />
-n subjobs Number of subjobs to run.<br />
-rflex file File listing protein residues to be treated flexibly. The residues are specified<br />
in the format chain:residue.<br />
-v Print version information and exit<br />
The output Maestro structure file includes a number of properties, which are described in<br />
Table 10.2.<br />
Table 10.2. Output properties from a <strong>Prime</strong> MM-GBSA calculation.<br />
Property Description<br />
<strong>Prime</strong> Coulomb Coulomb energy of the complex<br />
<strong>Prime</strong> Covalent Covalent binding energy of the complex<br />
<strong>Prime</strong> vdW Van der Waals energy of the complex<br />
<strong>Prime</strong> Solv SA Solvation surface area of the complex<br />
<strong>Prime</strong> Solv GB Solvation energy of the complex<br />
<strong>Prime</strong> Energy <strong>Prime</strong> energy of the complex<br />
<strong>Prime</strong> MMGBSA<br />
Complex Energy<br />
<strong>Prime</strong> MMGBSA Ligand<br />
Energy<br />
<strong>Prime</strong> MMGBSA<br />
Receptor Energy<br />
MMGBSA energy of the complex<br />
MMGBSA energy of the free ligand<br />
MMGBSA energy of the uncomplexed receptor<br />
<strong>Prime</strong> 2.1 <strong>User</strong> <strong>Manual</strong> 83