- Page 2 and 3:
Advances in Food Mycology
- Page 4 and 5:
Advances in Food Mycology Edited by
- Page 6 and 7:
FOREWORD This book represents the P
- Page 8 and 9:
CONTENTS Foreword .................
- Page 10 and 11:
Contents ix Mycobiota, mycotoxigeni
- Page 12 and 13:
CONTRIBUTORS M. Lourdes Abarca, Dep
- Page 14 and 15:
Contributors xiii Dilek Heperkan, I
- Page 16 and 17:
Section 1. Understanding the fungi
- Page 18 and 19:
4 Jens C. Frisvad et al. Furthermor
- Page 20 and 21:
6 Jens C. Frisvad et al. Table 1. M
- Page 22 and 23:
8 Jens C. Frisvad et al. Major sour
- Page 24 and 25:
10 Jens C. Frisvad et al. Major sou
- Page 26 and 27:
12 Jens C. Frisvad et al. 3.4. Cycl
- Page 28 and 29:
14 Jens C. Frisvad et al. and poult
- Page 30 and 31:
16 Jens C. Frisvad et al. 3.9. Zear
- Page 32 and 33:
18 Jens C. Frisvad et al. 4.5. Myco
- Page 34 and 35:
20 Jens C. Frisvad et al. 4.9. Peni
- Page 36 and 37:
22 Jens C. Frisvad et al. Major sou
- Page 38 and 39:
24 Jens C. Frisvad et al. only occu
- Page 40 and 41:
26 Jens C. Frisvad et al. 1977, Myc
- Page 42 and 43:
28 Jens C. Frisvad et al. Kamyar, M
- Page 44 and 45:
30 Jens C. Frisvad et al. Pitt, J.
- Page 46 and 47:
RECOMMENDATIONS CONCERNING THE CHRO
- Page 48 and 49: Recommendations Concerning the Chro
- Page 50 and 51: Recommendations Concerning the Chro
- Page 52 and 53: Recommendations Concerning the Chro
- Page 54 and 55: Recommendations Concerning the Chro
- Page 56 and 57: Recommendations Concerning the Chro
- Page 58 and 59: Recommendations Concerning the Chro
- Page 60 and 61: Section 2. Media and method develop
- Page 62 and 63: 50 M. H. Taniwaki et al. the counti
- Page 64 and 65: 52 M. H. Taniwaki et al. Table 1. O
- Page 66 and 67: 54 M. H. Taniwaki et al. 60 column
- Page 68 and 69: 56 M. H. Taniwaki et al. 3.2. Growt
- Page 70 and 71: 58 M. H. Taniwaki et al. 3.4. Estim
- Page 72 and 73: 60 M. H. Taniwaki et al. lowest; ab
- Page 74 and 75: 62 M. H. Taniwaki et al. 4. DISCUSS
- Page 76 and 77: 64 M. H. Taniwaki et al. viable cou
- Page 78 and 79: 66 M. H. Taniwaki et al. Brancato,
- Page 80 and 81: EVALUATION OF MOLECULAR METHODS FOR
- Page 82 and 83: Evaluation of Molecular Methods for
- Page 84 and 85: Evaluation of Molecular Methods for
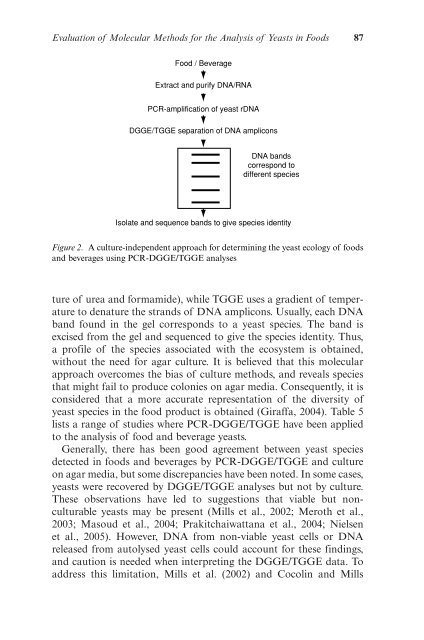
- Page 86 and 87: Evaluation of Molecular Methods for
- Page 88 and 89: Evaluation of Molecular Methods for
- Page 90 and 91: Evaluation of Molecular Methods for
- Page 92 and 93: Evaluation of Molecular Methods for
- Page 94 and 95: Evaluation of Molecular Methods for
- Page 96 and 97: Evaluation of Molecular Methods for
- Page 100 and 101: Evaluation of Molecular Methods for
- Page 102 and 103: Evaluation of Molecular Methods for
- Page 104 and 105: Evaluation of Molecular Methods for
- Page 106 and 107: Evaluation of Molecular Methods for
- Page 108 and 109: Evaluation of Molecular Methods for
- Page 110 and 111: Evaluation of Molecular Methods for
- Page 112 and 113: Evaluation of Molecular Methods for
- Page 114 and 115: Evaluation of Molecular Methods for
- Page 116 and 117: Evaluation of Molecular Methods for
- Page 118 and 119: STANDARDIZATION OF METHODS FOR DETE
- Page 120 and 121: Standardization of Methods for Dete
- Page 122 and 123: Standardization of Methods for Dete
- Page 124 and 125: ECOPHYSIOLOGY OF FUMONISIN PRODUCER
- Page 126 and 127: Ecophysiology of Fumonisin Producer
- Page 128 and 129: Ecophysiology of Fumonisin Producer
- Page 130 and 131: Ecophysiology of Fumonisin Producer
- Page 132 and 133: ECOPHYSIOLOGY OF FUSARIUM CULMORUM
- Page 134 and 135: Ecophysiology of Fusarium culmorum
- Page 136 and 137: Ecophysiology of Fusarium culmorum
- Page 138 and 139: Ecophysiology of Fusarium culmorum
- Page 140 and 141: Ecophysiology of Fusarium culmorum
- Page 142 and 143: Ecophysiology of Fusarium culmorum
- Page 144 and 145: Ecophysiology of Fusarium culmorum
- Page 146 and 147: FOOD-BORNE FUNGI IN FRUIT AND CEREA
- Page 148 and 149:
Fungi and Mycotoxins in Fruit and C
- Page 150 and 151:
Fungi and Mycotoxins in Fruit and C
- Page 152 and 153:
Fungi and Mycotoxins in Fruit and C
- Page 154 and 155:
Fungi and Mycotoxins in Fruit and C
- Page 156 and 157:
Fungi and Mycotoxins in Fruit and C
- Page 158 and 159:
Fungi and Mycotoxins in Fruit and C
- Page 160 and 161:
Fungi and Mycotoxins in Fruit and C
- Page 162 and 163:
BLACK ASPERGILLUS SPECIES IN AUSTRA
- Page 164 and 165:
Black Aspergillus Species in Austra
- Page 166 and 167:
Black Aspergillus Species in Austra
- Page 168 and 169:
Black Aspergillus Species in Austra
- Page 170 and 171:
Black Aspergillus Species in Austra
- Page 172 and 173:
Black Aspergillus Species in Austra
- Page 174 and 175:
Black Aspergillus Species in Austra
- Page 176 and 177:
Black Aspergillus Species in Austra
- Page 178 and 179:
Black Aspergillus Species in Austra
- Page 180 and 181:
Black Aspergillus Species in Austra
- Page 182 and 183:
174 Marta Bau et al. sporadically i
- Page 184 and 185:
176 Marta Bau et al. Table 1. Occur
- Page 186 and 187:
178 Marta Bau et al. Table 2. Ochra
- Page 188 and 189:
FUNGI PRODUCING OCHRATOXIN IN DRIED
- Page 190 and 191:
Fungi Producing Ochratoxin in Dried
- Page 192 and 193:
Fungi Producing Ochratoxin in Dried
- Page 194 and 195:
Fungi Producing Ochratoxin in Dried
- Page 196 and 197:
AN UPDATE ON OCHRATOXIGENIC FUNGI A
- Page 198 and 199:
An Update on Ochratoxigenic Fungi a
- Page 200 and 201:
An Update on Ochratoxigenic Fungi a
- Page 202 and 203:
An Update on Ochratoxigenic Fungi a
- Page 204 and 205:
An Update on Ochratoxigenic Fungi a
- Page 206 and 207:
An Update on Ochratoxigenic Fungi a
- Page 208 and 209:
An Update on Ochratoxigenic Fungi a
- Page 210 and 211:
MYCOBIOTA, MYCOTOXIGENIC FUNGI, AND
- Page 212 and 213:
Mycobiota and Citrinin in Black Oli
- Page 214 and 215:
Mycobiota and Citrinin in Black Oli
- Page 216 and 217:
Mycobiota and Citrinin in Black Oli
- Page 218 and 219:
BYSSOCHLAMYS: SIGNIFICANCE OF HEAT
- Page 220 and 221:
Byssochlamys Heat Resistance and My
- Page 222 and 223:
Byssochlamys Heat Resistance and My
- Page 224 and 225:
Byssochlamys Heat Resistance and My
- Page 226 and 227:
Byssochlamys Heat Resistance and My
- Page 228 and 229:
Byssochlamys Heat Resistance and My
- Page 230 and 231:
Byssochlamys Heat Resistance and My
- Page 232 and 233:
EFFECT OF WATER ACTIVITY AND TEMPER
- Page 234 and 235:
Aflatoxin and CPA Production in Pea
- Page 236 and 237:
Aflatoxin and CPA Production in Pea
- Page 238 and 239:
Aflatoxin and CPA Production in Pea
- Page 240 and 241:
Aflatoxin and CPA Production in Pea
- Page 242 and 243:
Aflatoxin and CPA Production in Pea
- Page 244 and 245:
INACTIVATION OF FRUIT SPOILAGE YEAS
- Page 246 and 247:
High Pressure Inactivation of Fungi
- Page 248 and 249:
High Pressure Inactivation of Fungi
- Page 250 and 251:
High Pressure Inactivation of Fungi
- Page 252 and 253:
ACTIVATION OF ASCOSPORES BY NOVEL F
- Page 254 and 255:
Activation of Ascospores by Novel F
- Page 256 and 257:
Activation of Ascospores by Novel F
- Page 258 and 259:
Activation of Ascospores by Novel F
- Page 260 and 261:
Activation of Ascospores by Novel F
- Page 262 and 263:
Activation of Ascospores by Novel F
- Page 264 and 265:
Activation of Ascospores by Novel F
- Page 266 and 267:
MIXTURES OF NATURAL AND SYNTHETIC A
- Page 268 and 269:
Mixtures of Natural and Synthetic A
- Page 270 and 271:
Mixtures of Natural and Synthetic A
- Page 272 and 273:
Mixtures of Natural and Synthetic A
- Page 274 and 275:
Mixtures of Natural and Synthetic A
- Page 276 and 277:
Mixtures of Natural and Synthetic A
- Page 278 and 279:
Mixtures of Natural and Synthetic A
- Page 280 and 281:
Mixtures of Natural and Synthetic A
- Page 282 and 283:
Mixtures of Natural and Synthetic A
- Page 284 and 285:
Mixtures of Natural and Synthetic A
- Page 286 and 287:
Mixtures of Natural and Synthetic A
- Page 288 and 289:
Mixtures of Natural and Synthetic A
- Page 290 and 291:
Mixtures of Natural and Synthetic A
- Page 292 and 293:
PROBABILISTIC MODELLING OF ASPERGIL
- Page 294 and 295:
Probabilistic Modelling of Aspergil
- Page 296 and 297:
Probabilistic Modelling of Aspergil
- Page 298 and 299:
Probabilistic Modelling of Aspergil
- Page 300 and 301:
Probabilistic Modelling of Aspergil
- Page 302 and 303:
Probabilistic Modelling of Aspergil
- Page 304 and 305:
Probabilistic Modelling of Aspergil
- Page 306 and 307:
Probabilistic Modelling of Aspergil
- Page 308 and 309:
Probabilistic Modelling of Aspergil
- Page 310 and 311:
Probabilistic Modelling of Aspergil
- Page 312 and 313:
ANTIFUNGAL ACTIVITY OF SOURDOUGH BR
- Page 314 and 315:
Antifungal Activity of Sourdough Br
- Page 316 and 317:
Antifungal Activity of Sourdough Br
- Page 318 and 319:
Antifungal Activity of Sourdough Br
- Page 320 and 321:
Antifungal Activity of Sourdough Br
- Page 322 and 323:
PREVENTION OF OCHRATOXIN A IN CEREA
- Page 324 and 325:
Prevention of Ochratoxin A in Cerea
- Page 326 and 327:
Prevention of Ochratoxin A in Cerea
- Page 328 and 329:
Prevention of Ochratoxin A in Cerea
- Page 330 and 331:
Prevention of Ochratoxin A in Cerea
- Page 332 and 333:
Prevention of Ochratoxin A in Cerea
- Page 334 and 335:
Prevention of Ochratoxin A in Cerea
- Page 336 and 337:
Prevention of Ochratoxin A in Cerea
- Page 338 and 339:
Prevention of Ochratoxin A in Cerea
- Page 340 and 341:
Prevention of Ochratoxin A in Cerea
- Page 342 and 343:
Prevention of Ochratoxin A in Cerea
- Page 344 and 345:
Prevention of Ochratoxin A in Cerea
- Page 346 and 347:
Prevention of Ochratoxin A in Cerea
- Page 348 and 349:
RECOMMENDED METHODS FOR FOOD MYCOLO
- Page 350 and 351:
Recommended Methods for Food Mycolo
- Page 352 and 353:
Recommended Methods for Food Mycolo
- Page 354 and 355:
APPENDIX 1 - MEDIA All media are st
- Page 356 and 357:
Appendix 1 - Media 351 CYA (1): Cza
- Page 358 and 359:
Appendix 1 - Media 353 note that th
- Page 360 and 361:
Appendix 1 - Media 355 PCA: Potato
- Page 362 and 363:
Appendix 1 - Media 357 Pitt, J. I.,
- Page 364 and 365:
Appendix 2 - International Commissi
- Page 366 and 367:
362 Index Antifungal agents (Contin
- Page 368 and 369:
364 Index Cellulase, from F. culmor
- Page 370 and 371:
366 Index Fusarenon X, 15 Fusarin C
- Page 372 and 373:
368 Index Mycophenolic acid, 18, 21
- Page 374 and 375:
370 Index Phaffia, 74 Phoma tenuazo